








生物技术通报 ›› 2023, Vol. 39 ›› Issue (9): 49-57.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0106
收稿日期:2023-02-13
出版日期:2023-09-26
发布日期:2023-10-24
通讯作者:
王猛,男,研究员,博士生导师,研究方向:合成生物学和高通量自动化等;E-mail: wangmeng@tib.cas.cn;作者简介:刘佳慧,女,硕士研究生,研究方向:生物与医药;E-mail: liujiah@tib.cas.cn
基金资助:
LIU Jia-hui1,2( ), LIU Ye2, HUA Er-bing1(
), LIU Ye2, HUA Er-bing1( ), WANG Meng2(
), WANG Meng2( )
)
Received:2023-02-13
Published:2023-09-26
Online:2023-10-24
摘要:
碱基编辑是一种新兴的基因组编辑技术,具有不产生双键断裂、不依赖同源重组且不需要添加外源模板的优势,在真核及原核生物中得到了广泛的开发与应用。为了进一步扩展碱基编辑技术在谷氨酸棒杆菌中的基因组覆盖范围,本研究将3种PAM限制较为宽松的新型Cas9突变体应用于胞嘧啶碱基编辑工具中,分别为近乎PAMless的SpRY突变体(NRN>NYN PAM)、SpG突变体(NGN PAM),以及ScCas9++蛋白(NNG PAM),实现对碱基编辑工具的PAM拓展。结合SpRY突变体的碱基编辑系统展示出了更宽松的PAM识别,除对CAT、CAC、TAA PAM的位点完全没有编辑外,对其他NRN种类的PAM位点均出现了不同程度的识别,但整体编辑效率低,难以推广应用;结合SpG突变体的碱基编辑系统可实现对所有NGN种类 PAM位点的编辑,且编辑效率优于SpRY突变体,但对NGG PAM位点的编辑,相比原始Cas9蛋白,编辑效率下降9.3%-55.9%;结合ScCas9++蛋白的碱基编辑系统,除对TCG、CTG PAM的基因组位点没有编辑外,可实现对其他测试NNG PAM的基因组位点编辑,大部分位点基因组编辑效率均较高,最高可达100%。本研究的开展不仅有助于碱基编辑工具在谷氨酸棒杆菌中覆盖更多的基因组位点,同时也为其他基于CRISPR/Cas系统的基因组编辑工具的PAM拓展提供有力的参考。
刘佳慧, 刘叶, 花尔并, 王猛. 谷氨酸棒杆菌中胞嘧啶碱基编辑工具的PAM拓展[J]. 生物技术通报, 2023, 39(9): 49-57.
LIU Jia-hui, LIU Ye, HUA Er-bing, WANG Meng. PAM Extension of Cytosine Base Editing Tool in Corynebacterium glutamicum[J]. Biotechnology Bulletin, 2023, 39(9): 49-57.
| Name | Source | |
|---|---|---|
| Plasmid | pXMJ19TS | Lab stock |
| pnCas9(D10A)-AIDTS | [15] | |
| pgRNA-ccdB | [15] | |
| pnCas9-Sc++(D10A)-AID TS | This study | |
| pnCas9-SpG(D10A)-AID TS | This study | |
| pnCas9-SpRY(D10A)-AID TS | This study | |
| Strain | Escherichia coli DH5α | Lab stock |
| C. glutamicum ATCC 13032 | Lab stock | |
表1 本文用到的质粒与菌株
Table 1 Plasmids and strains used in this study
| Name | Source | |
|---|---|---|
| Plasmid | pXMJ19TS | Lab stock |
| pnCas9(D10A)-AIDTS | [15] | |
| pgRNA-ccdB | [15] | |
| pnCas9-Sc++(D10A)-AID TS | This study | |
| pnCas9-SpG(D10A)-AID TS | This study | |
| pnCas9-SpRY(D10A)-AID TS | This study | |
| Strain | Escherichia coli DH5α | Lab stock |
| C. glutamicum ATCC 13032 | Lab stock | |
| Primer name | Sequence(5'-3') |
|---|---|
| ScD10A-1F | TAAGCTTAAAGGAGTTGAGAATGGAGAAGAAATACTCTATCGGCC |
| ScD10A-1R | GGAAGTCCAGGATGGTCTTGCCGGACTGCT |
| ScD10A-2F | CAAGACCATCCTGGACTTCCTGAAGTCCGATG |
| ScD10A-2R | ACCTTGCGTTTCTTCTTTGGATCGCCGCCCAACTGAGAGA |
| ScD10A-3F | CCAAAGAAGAAACGCAAGGTCG |
| S-3R | CCGCCGCCAAGTGATTCTTAG |
| ScD10A-4F | CTAAGAATCACTTGGCGGCGG |
| ScD10A-4R | TCTCAACTCCTTTAAGCTTAATTAA |
| SpG-1F | TAAGCTTAAAGGAGTTGAGAATGGAT |
| SpG-1R | AAAACCACCATATTTTTTTGGATCC |
| SpG-2F | CAAAAAAATATGGTGGTTTTCTGTGGCCAACGGTAGCTTAT |
| SpG-2R | TCTAAAACTTCTTTTGTAGAGCGATACTGTTTACGATCAATTGTT |
| S-3F | TCTACAAAAGAAGTTTTAGATGCCAC |
| SpG-4F | CTAAGAATCACTTGGCGGC |
| SpG-4R | TCTCAACTCCTTTAAGCTTAATTAATTCT |
| SpRY-1F | AGCGGAACGCACTCGTCTCA |
| SpRY-1R | AATTGACTCCTTGGAGAATCCGC |
| SpRY-2F | GATTCTCCAAGGAGTCAATTCGCCCAAAAAGAAATTCGGACA |
| SpRY-2R | TCTAAAACTTCTTTTGTAGAGCGATACTGTTTTGGATCAATTGTT |
| SpRY-4F | CTAAGAATCACTTGGCGGCGGGTG |
| SpRY-4R | TGAGACGAGTGCGTTCCGCTGTCTC |
表2 本文质粒载体构建用到的引物
Table 2 Primers for plasmids construction used in this study
| Primer name | Sequence(5'-3') |
|---|---|
| ScD10A-1F | TAAGCTTAAAGGAGTTGAGAATGGAGAAGAAATACTCTATCGGCC |
| ScD10A-1R | GGAAGTCCAGGATGGTCTTGCCGGACTGCT |
| ScD10A-2F | CAAGACCATCCTGGACTTCCTGAAGTCCGATG |
| ScD10A-2R | ACCTTGCGTTTCTTCTTTGGATCGCCGCCCAACTGAGAGA |
| ScD10A-3F | CCAAAGAAGAAACGCAAGGTCG |
| S-3R | CCGCCGCCAAGTGATTCTTAG |
| ScD10A-4F | CTAAGAATCACTTGGCGGCGG |
| ScD10A-4R | TCTCAACTCCTTTAAGCTTAATTAA |
| SpG-1F | TAAGCTTAAAGGAGTTGAGAATGGAT |
| SpG-1R | AAAACCACCATATTTTTTTGGATCC |
| SpG-2F | CAAAAAAATATGGTGGTTTTCTGTGGCCAACGGTAGCTTAT |
| SpG-2R | TCTAAAACTTCTTTTGTAGAGCGATACTGTTTACGATCAATTGTT |
| S-3F | TCTACAAAAGAAGTTTTAGATGCCAC |
| SpG-4F | CTAAGAATCACTTGGCGGC |
| SpG-4R | TCTCAACTCCTTTAAGCTTAATTAATTCT |
| SpRY-1F | AGCGGAACGCACTCGTCTCA |
| SpRY-1R | AATTGACTCCTTGGAGAATCCGC |
| SpRY-2F | GATTCTCCAAGGAGTCAATTCGCCCAAAAAGAAATTCGGACA |
| SpRY-2R | TCTAAAACTTCTTTTGTAGAGCGATACTGTTTTGGATCAATTGTT |
| SpRY-4F | CTAAGAATCACTTGGCGGCGGGTG |
| SpRY-4R | TGAGACGAGTGCGTTCCGCTGTCTC |
| gRNA ID | Sequence(5'-3') | PAM |
|---|---|---|
| Cgl0871-AAG | AACCAAAGAGATGGATTTGG | AAG |
| Cgl1928-ATG | AGTCCCTTCCGCGTCTGCGC | ATG |
| Cgl1738-ACG | GCCCACATGACAAAATGCTC | ACG |
| Cgl2291-AGG | TCAGCGCAACGTGGAACTTG | AGG |
| Cgl1408-TAG | CAGTCAAACGCTCGAGAAAC | TAG |
| Cgl2146-TTG | CCAGCGTCCGCGCAAATTTC | TTG |
| Cgl17576-TCG | CACTCAGCTTCGTGCTGAAC | TCG |
| Cgl2965-TGG | ACAACCCCTGGCTCAGATGG | TGG |
| Cgl0863-CAG | TTCCACTTGGTGGACAGCAG | CAG |
| Cgl1806-CTG | CGACATAAACAAGGAGGCAC | CTG |
| Cgl2111-CCG | CGTCGAACACCTCTATCCCA | CCG |
| Cgl0179-CGG | GCACCACTGCATCTACCTGG | CGG |
| Cgl0037-GAG | TCAAGGAACATCCACCGTTC | GAG |
| Cgl2426-GTG | CACACTAGGCGCGAACTATC | GTG |
| Cgl2608-GCG | TACCACTGACCGATGCTTGC | GCG |
| Cgl0170-GGG | GTGCCTGCGATGACAAATGG | GGG |
| Cgl1467-AGA | AATCCACGTGGTAACCAGGT | AGA |
| Cgl2044-AGT | ACACATACCCCTTGCCAGAT | AGT |
| Cgl3043-AGC | CCCGAAACAAAGAGGCCATC | AGC |
| Cgl2647-TGA | CTCACAGAGTGGGCAGGCAC | TGA |
| Cgl1820-TGC | CGCTATGCGCTAGCGGTAGA | TGC |
| Cgl2711-CGA | GCCGCAGCAATTATCTCCAC | CGA |
| Cgl0698-CGT | GCACCGTGGCAGTGAGTGGC | CGT |
| Cgl2647-CGC | TCCCAGAATTACACCAGGAG | CGC |
| Cgl1890-GGA | TCCCAGCGTGCACAATACGT | GGA |
| Cgl1839-GGT | ACCACAGGTGAGACAGTTCA | GGT |
| Cgl1535-GGC | CCCCACACTTTCTCCACGAT | GGC |
| Cgl2009-AAA | CCCACCATGTCCACTATCTC | AAA |
| Cgl1008-AAT | CCCAATGTGTCCTATAGCAC | AAT |
| Cgl0170-AAC | ACCTCACCACCATCGACGAC | AAC |
| Cgl1011-TAA | CACCCCTAGAGTGAGGTGGG | TAA |
| Cgl2711-TAT | CTGGCCATTGATGCATCGGA | TAT |
| Cgl2035-TAC | ATCACACGTGTGCCGAAAAA | TAC |
| Cgl0379-CAA | AACCTCTTTGGCAACCACGA | CAA |
| Cgl2111-CAC | TCCTGCTGCTTCGCTGCTGC | CAC |
| Cgl2148-GAA | ACCAGGCACCAGCTTTCGGT | GAA |
| Cgl1108-GAT | GACCAATTTGCTTGGCCTTC | GAT |
| Cgl1239-GAC | GCACTGCATTCCGCAAACCC | GAC |
| Cgl2475-TGT | GCTCCATTCAGACCATGGAA | TGT |
| Cgl1925-CAT | CCTCTCCCAGGCACTTGTCG | CAT |
| Cgl0871-AAG | AACCAAAGAGATGGATTTGG | AAG |
| Cgl0982-AAG | CCAGGTTGCCGATGATTCTC | AAG |
| Cgl1928-ATG | AGTCCCTTCCGCGTCTGCGC | ATG |
| Cgl1738-ACG | GCCCACATGACAAAATGCTC | ACG |
| Cgl2291-AGG | TCAGCGCAACGTGGAACTTG | AGG |
| Cgl1408-TAG | CAGTCAAACGCTCGAGAAAC | TAG |
| Cgl2146-TTG | CCAGCGTCCGCGCAAATTTC | TTG |
表3 本文所用到的gRNA
Table 3 gRNAs used in this study
| gRNA ID | Sequence(5'-3') | PAM |
|---|---|---|
| Cgl0871-AAG | AACCAAAGAGATGGATTTGG | AAG |
| Cgl1928-ATG | AGTCCCTTCCGCGTCTGCGC | ATG |
| Cgl1738-ACG | GCCCACATGACAAAATGCTC | ACG |
| Cgl2291-AGG | TCAGCGCAACGTGGAACTTG | AGG |
| Cgl1408-TAG | CAGTCAAACGCTCGAGAAAC | TAG |
| Cgl2146-TTG | CCAGCGTCCGCGCAAATTTC | TTG |
| Cgl17576-TCG | CACTCAGCTTCGTGCTGAAC | TCG |
| Cgl2965-TGG | ACAACCCCTGGCTCAGATGG | TGG |
| Cgl0863-CAG | TTCCACTTGGTGGACAGCAG | CAG |
| Cgl1806-CTG | CGACATAAACAAGGAGGCAC | CTG |
| Cgl2111-CCG | CGTCGAACACCTCTATCCCA | CCG |
| Cgl0179-CGG | GCACCACTGCATCTACCTGG | CGG |
| Cgl0037-GAG | TCAAGGAACATCCACCGTTC | GAG |
| Cgl2426-GTG | CACACTAGGCGCGAACTATC | GTG |
| Cgl2608-GCG | TACCACTGACCGATGCTTGC | GCG |
| Cgl0170-GGG | GTGCCTGCGATGACAAATGG | GGG |
| Cgl1467-AGA | AATCCACGTGGTAACCAGGT | AGA |
| Cgl2044-AGT | ACACATACCCCTTGCCAGAT | AGT |
| Cgl3043-AGC | CCCGAAACAAAGAGGCCATC | AGC |
| Cgl2647-TGA | CTCACAGAGTGGGCAGGCAC | TGA |
| Cgl1820-TGC | CGCTATGCGCTAGCGGTAGA | TGC |
| Cgl2711-CGA | GCCGCAGCAATTATCTCCAC | CGA |
| Cgl0698-CGT | GCACCGTGGCAGTGAGTGGC | CGT |
| Cgl2647-CGC | TCCCAGAATTACACCAGGAG | CGC |
| Cgl1890-GGA | TCCCAGCGTGCACAATACGT | GGA |
| Cgl1839-GGT | ACCACAGGTGAGACAGTTCA | GGT |
| Cgl1535-GGC | CCCCACACTTTCTCCACGAT | GGC |
| Cgl2009-AAA | CCCACCATGTCCACTATCTC | AAA |
| Cgl1008-AAT | CCCAATGTGTCCTATAGCAC | AAT |
| Cgl0170-AAC | ACCTCACCACCATCGACGAC | AAC |
| Cgl1011-TAA | CACCCCTAGAGTGAGGTGGG | TAA |
| Cgl2711-TAT | CTGGCCATTGATGCATCGGA | TAT |
| Cgl2035-TAC | ATCACACGTGTGCCGAAAAA | TAC |
| Cgl0379-CAA | AACCTCTTTGGCAACCACGA | CAA |
| Cgl2111-CAC | TCCTGCTGCTTCGCTGCTGC | CAC |
| Cgl2148-GAA | ACCAGGCACCAGCTTTCGGT | GAA |
| Cgl1108-GAT | GACCAATTTGCTTGGCCTTC | GAT |
| Cgl1239-GAC | GCACTGCATTCCGCAAACCC | GAC |
| Cgl2475-TGT | GCTCCATTCAGACCATGGAA | TGT |
| Cgl1925-CAT | CCTCTCCCAGGCACTTGTCG | CAT |
| Cgl0871-AAG | AACCAAAGAGATGGATTTGG | AAG |
| Cgl0982-AAG | CCAGGTTGCCGATGATTCTC | AAG |
| Cgl1928-ATG | AGTCCCTTCCGCGTCTGCGC | ATG |
| Cgl1738-ACG | GCCCACATGACAAAATGCTC | ACG |
| Cgl2291-AGG | TCAGCGCAACGTGGAACTTG | AGG |
| Cgl1408-TAG | CAGTCAAACGCTCGAGAAAC | TAG |
| Cgl2146-TTG | CCAGCGTCCGCGCAAATTTC | TTG |
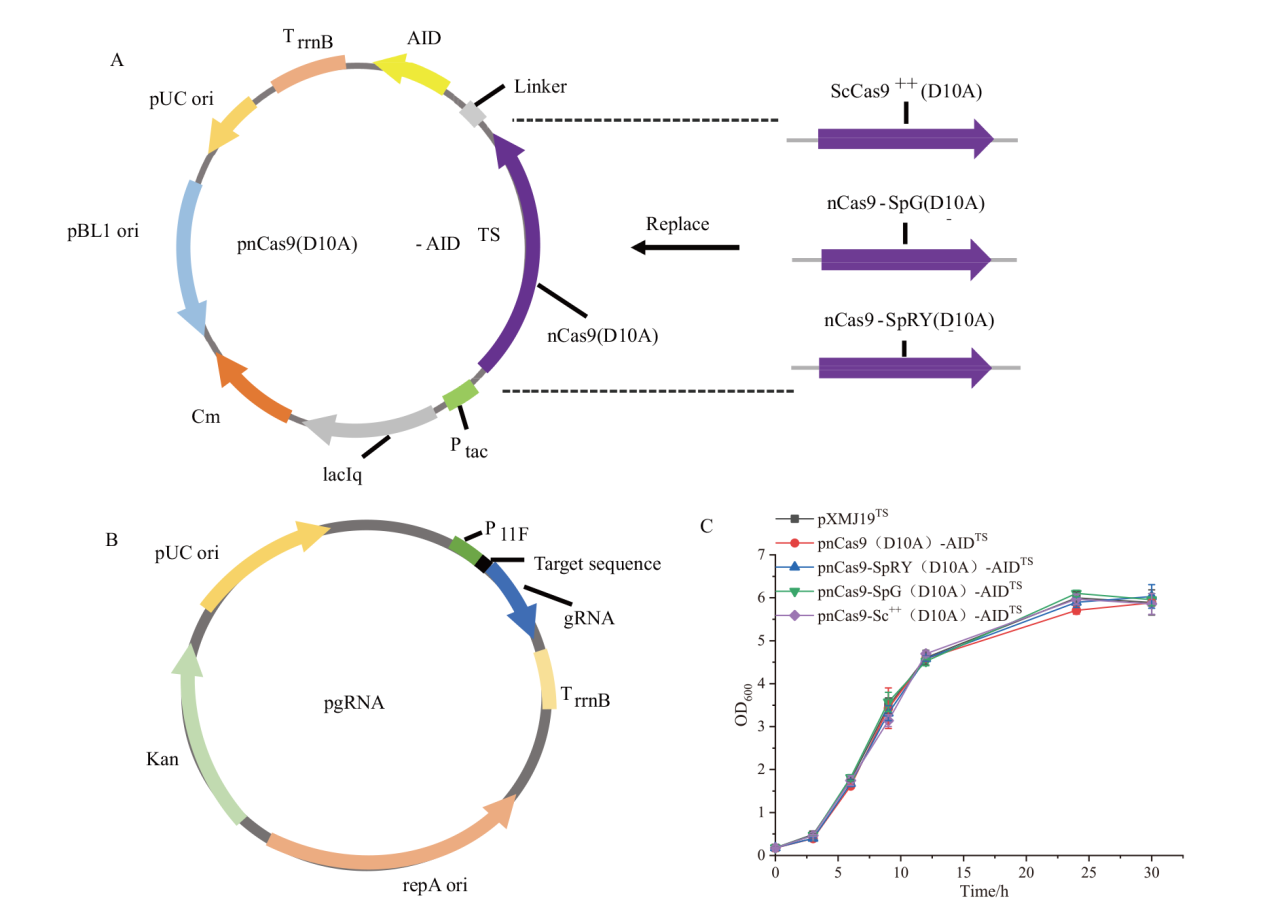
图1 碱基编辑质粒的构建 A:含不同Cas9突变体碱基编辑质粒的构建(Ptac:Tac 启动子;TrrnB:rrnB 终止子;Cm:氯霉素);B:gRNA表达质粒的构建(P11F:谷氨酸棒杆菌中cspB 启动子的衍生[22]:Kan:卡那霉素);C:结合不同Cas9蛋白的胞嘧啶碱基编辑工具对菌株生长的影响(平行样品数量n=3)
Fig. 1 Plasmid construction for base editing A: Plasmid construction for base editing with different Cas9 mutants(Ptac: Tac promoter; TrrnB: rrnB terminator; Cm: chloroamphenicol). B: Plasmid construction for gRNA expression(P11F: a derivative of cspB promoter from C. glutamicum[22]; Kan:Kanamycin). C: Effect of cytosine base editing tools combined with different Cas9 proteins on strain growth(Number of replicates n=3)

图2 nCas9-SpRY(D10A)-AID的碱基编辑结果 A: NGN PAM的编辑效率;B:NAN PAM的编辑效率。平行样品数量n=3,* P≤0.05,** P≤0.01,*** P≤0.001,**** P≤0.000 1,student’s two-tailed t-test,下同
Fig. 2 Base editing results of nCas9-SpRY(D10A)-AID A: Editing efficiency for NGN PAMs. B: Editing efficiency for NAN PAMs. Number of replicates n=3, * P≤0.05, ** P≤0.01, *** P≤0.001, **** P≤0.000 1, student’s two-tailed t-test, the same below
| [1] |
Wang Y, Liu Y, Zheng P, et al. Microbial base editing: a powerful emerging technology for microbial genome engineering[J]. Trends Biotechnol, 2021, 39(2): 165-180.
doi: 10.1016/j.tibtech.2020.06.010 pmid: 32680590 |
| [2] |
Komor AC, Kim YB, Packer MS, et al. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage[J]. Nature, 2016, 533(7603): 420-424.
doi: 10.1038/nature17946 |
| [3] | Nishida K, Arazoe T, Yachie N, et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems[J]. Science, 2016, 353(6305): eaaf8729. |
| [4] |
Gaudelli NM, Komor AC, Rees HA, et al. Programmable base editing of A·T to G·C in genomic DNA without DNA cleavage[J]. Nature, 2017, 551(7681): 464-471.
doi: 10.1038/nature24644 URL |
| [5] | Zhao DD, Li J, Li SW, et al. Publisher correction: Glycosylase base editors enable C-to-A and C-to-G base changes[J]. Nat Biotechnol, 2021, 39(1): 115. |
| [6] |
Deng C, Lv XQ, Li JH, et al. Development of a DNA double-strand break-free base editing tool in Corynebacterium glutamicum for genome editing and metabolic engineering[J]. Metab Eng Commun, 2020, 11: e00135.
doi: 10.1016/j.mec.2020.e00135 URL |
| [7] |
Anzalone AV, Randolph PB, Davis JR, et al. Search-and-replace genome editing without double-strand breaks or donor DNA[J]. Nature, 2019, 576(7785): 149-157.
doi: 10.1038/s41586-019-1711-4 |
| [8] |
Tong YJ, Jørgensen TS, Whitford CM, et al. A versatile genetic engineering toolkit for E. coli based on CRISPR-prime editing[J]. Nat Commun, 2021, 12: 5206.
doi: 10.1038/s41467-021-25541-3 |
| [9] |
Yang B, Yang L, Chen J. Development and application of base editors[J]. CRISPR J, 2019, 2(2): 91-104.
doi: 10.1089/crispr.2019.0001 pmid: 30998092 |
| [10] |
Anzalone AV, Koblan LW, Liu DR. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors[J]. Nat Biotechnol, 2020, 38(7): 824-844.
doi: 10.1038/s41587-020-0561-9 pmid: 32572269 |
| [11] |
Yu SY, Birkenshaw A, Thomson T, et al. Increasing the targeting scope of CRISPR base editing system beyond NGG[J]. CRISPR J, 2022, 5(2): 187-202.
doi: 10.1089/crispr.2021.0109 URL |
| [12] |
Walton RT, Christie KA, Whittaker MN, et al. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants[J]. Science, 2020, 368(6488): 290-296.
doi: 10.1126/science.aba8853 pmid: 32217751 |
| [13] |
Chatterjee P, Jakimo N, Lee J, et al. An engineered ScCas9 with broad PAM range and high specificity and activity[J]. Nat Biotechnol, 2020, 38(10): 1154-1158.
doi: 10.1038/s41587-020-0517-0 |
| [14] |
Becker J, Wittmann C. Bio-based production of chemicals, materials and fuels-Corynebacterium glutamicum as versatile cell factory[J]. Curr Opin Biotechnol, 2012, 23(4): 631-640.
doi: 10.1016/j.copbio.2011.11.012 URL |
| [15] |
Wang Y, Liu Y, Liu J, et al. MACBETH: Multiplex automated Corynebacterium glutamicum base editing method[J]. Metab Eng, 2018, 47: 200-210.
doi: S1096-7176(17)30417-2 pmid: 29580925 |
| [16] |
Wang Y, Liu Y, Li JW, et al. Expanding targeting scope, editing window, and base transition capability of base editing in Corynebacterium glutamicum[J]. Biotechnol Bioeng, 2019, 116(11): 3016-3029.
doi: 10.1002/bit.27121 pmid: 31317533 |
| [17] |
黄华媚, 白立宽, 刘叶, 等. BE3型胞嘧啶碱基编辑器在谷氨酸棒杆菌中的开发及应用[J]. 生物技术通报, 2020, 36(3): 95-101.
doi: 10.13560/j.cnki.biotech.bull.1985.2019-0956 |
|
Huang HM, Bai LK, Liu Y, et al. Development and application of BE3 cytidine base editor in Corynebacterium glutamicum[J]. Biotechnol Bull, 2020, 36(3): 95-101.
doi: 10.13560/j.cnki.biotech.bull.1985.2019-0956 |
|
| [18] | 李俊维, 刘叶, 王钰, 等. 谷氨酸棒杆菌碱基编辑的条件优化[J]. 生物工程学报, 2020, 36(1): 143-151. |
| Li JW, Liu Y, Wang Y, et al. Optimization of base editing in Cory-nebacterium glutamicum[J]. Chin J Biotechnol, 2020, 36(1): 143-151. | |
| [19] | 卢挥, 张启, 于思礼, 等. 谷氨酸棒杆菌中基于CRISPR/Cas9的多位点碱基编辑系统的优化[J]. 生物工程学报, 2022, 38(2): 780-795. |
| Lu H, Zhang Q, Yu SL, et al. Optimization of CRISPR/Cas9-based multiplex base editing in Corynebacterium glutamicum[J]. Chin J Biotechnol, 2022, 38(2): 780-795. | |
| [20] |
Ruan YL, Zhu LJ, Li Q. Improving the electro-transformation efficiency of Corynebacterium glutamicum by weakening its cell wall and increasing the cytoplasmic membrane fluidity[J]. Biotechnol Lett, 2015, 37(12): 2445-2452.
doi: 10.1007/s10529-015-1934-x URL |
| [21] |
Kluesner MG, Nedveck DA, Lahr WS, et al. EditR: a method to quantify base editing from Sanger sequencing[J]. CRISPR J, 2018, 1(3): 239-250.
doi: 10.1089/crispr.2018.0014 URL |
| [22] |
Peyret JL, Bayan N, Joliff G, et al. Characterization of the cspB gene encoding PS2, an ordered surface-layer protein in Corynebacterium glutamicum[J]. Mol Microbiol, 1993, 9(1): 97-109.
doi: 10.1111/j.1365-2958.1993.tb01672.x pmid: 8412676 |
| [23] |
Wang Y, Cheng HJ, Liu Y, et al. In-situ generation of large numbers of genetic combinations for metabolic reprogramming via CRISPR-guided base editing[J]. Nat Commun, 2021, 12: 678.
doi: 10.1038/s41467-021-21003-y pmid: 33514753 |
| [24] | Liu Y, Wang RY, Liu JH, et al. Base editor enables rational genome-scale functional screening for enhanced industrial phenotypes in Corynebacterium glutamicum[J]. Sci Adv, 2022, 8(35): eabq2157. |
| [25] |
Tian KR, Hong X, Guo MM, et al. Development of base editors for simultaneously editing multiple loci in Lactococcus lactis[J]. ACS Synth Biol, 2022, 11(11): 3644-3656.
doi: 10.1021/acssynbio.1c00561 URL |
| [1] | 薛宁, 王瑾, 李世新, 刘叶, 程海娇, 张玥, 毛雨丰, 王猛. 多基因同步调控结合高通量筛选构建高产L-苯丙氨酸的谷氨酸棒杆菌工程菌株[J]. 生物技术通报, 2023, 39(9): 268-280. |
| [2] | 卢振万, 李雪琪, 黄金光, 周焕斌. 利用胞嘧啶碱基编辑技术创制耐草甘膦水稻[J]. 生物技术通报, 2023, 39(2): 63-69. |
| [3] | 聂立斌, 易铃欣, 邓妍, 盛琦, 吴晓玉, 张斌. 途径工程改造谷氨酸棒杆菌产莽草酸[J]. 生物技术通报, 2022, 38(6): 93-102. |
| [4] | 胡孝林, 王良婷, 顾玮, 罗阳. 基于CRISPR/Cas系统的生物传感策略研究进展[J]. 生物技术通报, 2020, 36(3): 69-77. |
| [5] | 黄华媚, 白立宽, 刘叶, 李俊维, 王猛, 花尔并. BE3型胞嘧啶碱基编辑器在谷氨酸棒杆菌中的开发用[J]. 生物技术通报, 2020, 36(3): 95-101. |
| [6] | 聂志华, 朱蕾蕾. 生物素对发酵过程中MscCG外排L-谷氨酸的影响[J]. 生物技术通报, 2020, 36(10): 150-155. |
| [7] | 刘妮, 陆沁, 刘娟, 陈航. CRISPR/Cas系统最新研究进展[J]. 生物技术通报, 2017, 33(2): 53-58. |
| [8] | 徐德雨, 郑小梅, 赵晶, 郑平, 赵树欣. 谷氨酸棒杆菌天冬氨酸激酶G359D突变解除赖氨酸与苏氨酸协同抑制的研究[J]. 生物技术通报, 2017, 33(11): 143-152. |
| [9] | 石增秀 崔文璟 周丽 周哲敏. 谷氨酸棒杆菌L-天冬氨酸α-脱羧酶基因的克隆及重组酶性质研究[J]. 生物技术通报, 2013, 0(4): 110-115. |
| [10] | 罗玉常;窦文芳;张晓梅;史劲松;许正宏;. 谷氨酸棒杆菌ilvE基因的敲除对相关氨基酸合成的影响[J]. , 2012, 0(11): 185-191. |
| [11] | 黄云雁;黎明;刘萌;孙昕;周丽颖;路福平;. 赖氨酸-尸胺反向转运蛋白cadB基因谷氨酸棒杆菌表达载体的构建及转化[J]. , 2012, 0(08): 94-100. |
| [12] | 伍展红;郑穗平;. 谷氨酸棒杆菌ldh基因的敲除[J]. , 2012, 0(02): 107-111. |
| [13] | 李智涛;伍展红;郑穗平;. 谷氨酸棒杆菌aroⅡ、trpEGD基因的过量表达及其对色氨酸合成的影响[J]. , 2011, 0(05): 151-156. |
| [14] | 吕扬勇;伍展红;郑穗平;. 谷氨酸棒杆菌metX、dapA基因敲除对苏氨酸合成的影响[J]. , 2011, 0(01): 158-164. |
| [15] | 林琳;窦文芳;张晓梅;许泓瑜;许正宏;王正祥;. 直接利用糖质原料产L-丝氨酸谷氨酸棒杆菌glyA基因序列分析[J]. , 2008, 0(05): 176-180. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||