








生物技术通报 ›› 2022, Vol. 38 ›› Issue (8): 127-134.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1519
李秀青1( ), 胡子曜2, 雷建峰1, 代培红2, 刘超2, 邓嘉辉2, 刘敏2, 孙玲1, 刘晓东2, 李月2(
), 胡子曜2, 雷建峰1, 代培红2, 刘超2, 邓嘉辉2, 刘敏2, 孙玲1, 刘晓东2, 李月2( )
)
收稿日期:2021-12-07
出版日期:2022-08-26
发布日期:2022-09-14
作者简介:李秀青,男,硕士,研究方向:作物逆境分子生物学;E-mail: 基金资助:
LI Xiu-qing1( ), HU Zi-yao2, LEI Jian-feng1, DAI Pei-hong2, LIU Chao2, DENG Jia-hui2, LIU Min2, SUN Ling1, LIU Xiao-dong2, LI Yue2(
), HU Zi-yao2, LEI Jian-feng1, DAI Pei-hong2, LIU Chao2, DENG Jia-hui2, LIU Min2, SUN Ling1, LIU Xiao-dong2, LI Yue2( )
)
Received:2021-12-07
Published:2022-08-26
Online:2022-09-14
摘要:
初步探究陆地棉TIFY家族基因GhTIFY9在棉花黄萎病反应中的功能,为挖掘棉花抗病相关基因及棉花抗病育种奠定基础。通过转录组筛选、克隆获得一个TIFY家族基因GhTIFY9,利用生物信息学的方法分析该基因的理化性质,通过荧光定量PCR(real-time quantitative polymerase chain reaction,RT-qPCR)方法分析GhTIFY9在黄萎病诱导下的表达模式。同时构建该基因的VIGS载体,利用病毒诱导的基因沉默(virus-induced gene silencing,VIGS)技术对其在棉花抗黄萎病中的功能进行了初步探究。结果表明,以陆地棉TM-1的cDNA为模板克隆获得GhTIFY9,其开放阅读框(ORF)为594 bp,编码一个含197个氨基酸的碱性亲水性蛋白,相对分子质量为21.65 kD,定位于细胞核,属于TIFY亚家族。RT-qPCR分析结果表明GhTIFY9响应黄萎病菌诱导,抑制该基因表达后沉默植株对黄萎病菌的敏感性显著增强。GhTIFY9是棉花抗黄萎病反应的一个正向调控因子。
李秀青, 胡子曜, 雷建峰, 代培红, 刘超, 邓嘉辉, 刘敏, 孙玲, 刘晓东, 李月. 棉花黄萎病抗性相关基因GhTIFY9的克隆与功能分析[J]. 生物技术通报, 2022, 38(8): 127-134.
LI Xiu-qing, HU Zi-yao, LEI Jian-feng, DAI Pei-hong, LIU Chao, DENG Jia-hui, LIU Min, SUN Ling, LIU Xiao-dong, LI Yue. Cloning and Functional Analysis of Gene GhTIFY9 Related to Cotton Verticillium Wilt Resistance[J]. Biotechnology Bulletin, 2022, 38(8): 127-134.
| 基因 Gene | 开放阅读框ORF/bp | 氨基酸 Amino acid | 相对分子质量Relative molecular weight/kD | 理论等电点Theoretical isoelectric point | 平均疏水性Average hydrophobicity | 信号肽 Signal peptide | 亚细胞定位 Subcellular localization | 脂肪系数Fat coefficient | 跨膜结构域 Transmembrane domain |
|---|---|---|---|---|---|---|---|---|---|
| GhTIFY9 | 594 | 197 | 21.65 | 9.08 | -0.477 | 无 None | 细胞核 Nucleus | 76.63 | 无 None |
表1 GhTIFY9的生物信息学分析
Table 1 Bioinformatics analysis of GhTIFY9
| 基因 Gene | 开放阅读框ORF/bp | 氨基酸 Amino acid | 相对分子质量Relative molecular weight/kD | 理论等电点Theoretical isoelectric point | 平均疏水性Average hydrophobicity | 信号肽 Signal peptide | 亚细胞定位 Subcellular localization | 脂肪系数Fat coefficient | 跨膜结构域 Transmembrane domain |
|---|---|---|---|---|---|---|---|---|---|
| GhTIFY9 | 594 | 197 | 21.65 | 9.08 | -0.477 | 无 None | 细胞核 Nucleus | 76.63 | 无 None |
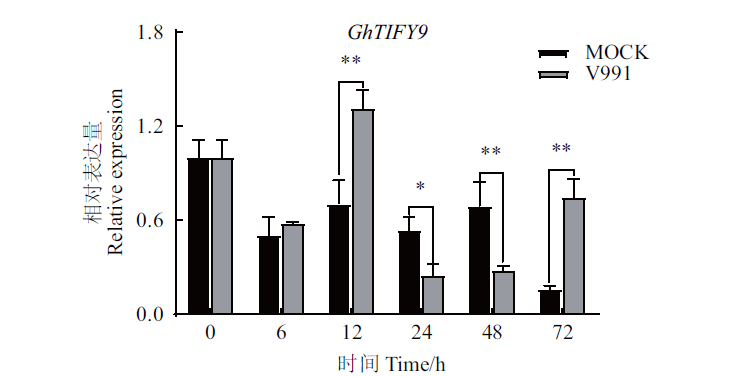
图3 黄萎病诱导下GhTIFY9的表达模式分析 *:在0.05水平上存在显著差异,**:在0.01水平上存在显著差异。下同
Fig.3 Analysis of the expression patterns of GhTIFY9 under the treatment of Verticillium dahliae *:Significant difference at the 0.05 level. **:Significant difference at the 0.01 level. The same as below
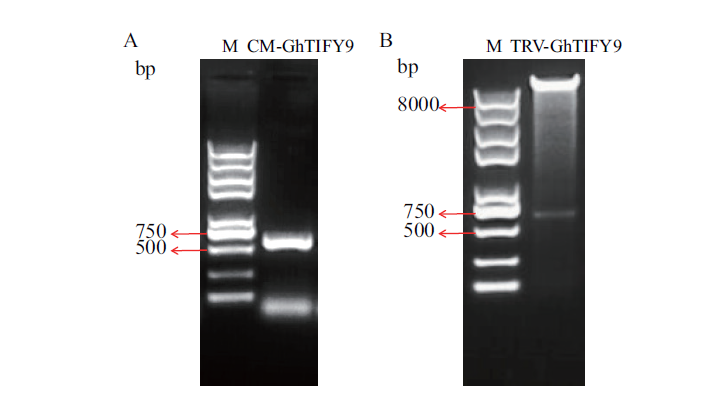
图4 GhTIFY9的VIGS载体构建 A:GhTIFY9目标片段的PCR扩增;B:TRV﹕GhTIFY9 VIGS载体的酶切鉴定。M:Trans 2K Plus II DNA marker
Fig. 4 VIGS vector construction of GhTIFY9 A:PCR amplification of target fragment of GhTIFY9. B:Identification of TRV﹕GhTIFY9 vectors by restriction enzyme digestion. M:Trans 2K Plus II DNA marker

图5 GhTIFY9的沉默效率检测 A:在棉花中沉默GhCLA1的表型;B:GhCLA1的沉默效率检测;C:GhTIFY9的沉默效率检测
Fig. 5 Silencing efficiency detection of GhTIFY9 gene A:The phenotype of silenced GhCLA1 in cotton. B:Silencing efficiency detection of GhCLA1. C:Silencing efficiency detection of GhTIFY9
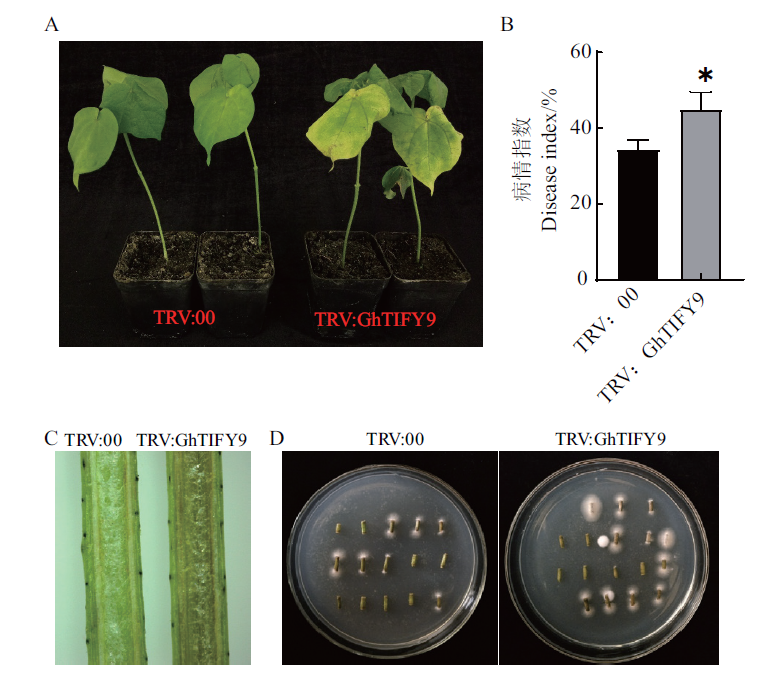
图6 GhTIFY9沉默植株的抗病性鉴定 A:GhTIFY9沉默后植株的表型分析;B:病情指数统计;C:剖杆鉴定;D:病原菌恢复培养
Fig. 6 Disease resistance identification of GhTIFY9 gene-silenced plants A:Phenotype analysis of plants after GhTIFY9 gene silencing. B:Disease index statistics. C:Identification by stem anatomy. D:Restoration culture of pathogenic bacteria
| [1] |
Li FG, Fan GY, Lu CR, et al. Genome sequence of cultivated Upland cotton(Gossypium hirsutum TM-1)provides insights into genome evolution[J]. Nat Biotechnol, 2015, 33(5):524-530.
doi: 10.1038/nbt.3208 URL |
| [2] |
Yang CL, Liang S, Wang HY, et al. Cotton major latex protein 28 functions as a positive regulator of the ethylene responsive factor 6 in defense against Verticillium dahliae[J]. Mol Plant, 2015, 8(3):399-411.
doi: 10.1016/j.molp.2014.11.023 URL |
| [3] |
Gong Q, Yang ZE, Wang XQ, et al. Salicylic acid-related cotton(Gossypium arboreum)ribosomal protein GaRPL18 contributes to resistance to Verticillium dahliae[J]. BMC Plant Biol, 2017, 17(1):59.
doi: 10.1186/s12870-017-1007-5 URL |
| [4] |
He X, Kang Y, Li WQ, et al. Genome-wide identification and functional analysis of the TIFY gene family in the response to multiple stresses in Brassica napus L[J]. BMC Genomics, 2020, 21(1):736.
doi: 10.1186/s12864-020-07128-2 URL |
| [5] |
Bai YH, Meng YJ, Huang DL, et al. Origin and evolutionary analysis of the plant-specific TIFY transcription factor family[J]. Genomics, 2011, 98(2):128-136.
doi: 10.1016/j.ygeno.2011.05.002 URL |
| [6] |
Nishii A, Takemura M, Fujita H, et al. Characterization of a novel gene encoding a putative single zinc-finger protein, ZIM, expressed during the reproductive phase in Arabidopsis thaliana[J]. Biosci Biotechnol Biochem, 2000, 64(7):1402-1409.
doi: 10.1271/bbb.64.1402 URL |
| [7] |
杨锐佳, 张中保, 吴忠义. 植物转录因子TIFY家族蛋白结构和功能的研究进展[J]. 生物技术通报, 2020, 36(12):121-128.
doi: 10.13560/j.cnki.biotech.bull.1985.2020-0307 |
| Yang RJ, Zhang ZB, Wu ZY. Progress of the structural and functional analysis of plant transcription factor TIFY protein family[J]. Biotechnol Bull, 2020, 36(12):121-128. | |
| [8] |
Meng L, Zhang T, Geng SS, et al. Comparative proteomics and metabolomics of JAZ7-mediated drought tolerance in Arabidopsis[J]. J Proteomics, 2019, 196:81-91.
doi: S1874-3919(19)30036-3 pmid: 30731210 |
| [9] |
Ebel C, BenFeki A, Hanin M, et al. Characterization of wheat(Tri-ticum aestivum)TIFY family and role of Triticum Durum TdTIFY11a in salt stress tolerance[J]. PLoS One, 2018, 13(7):e0200566.
doi: 10.1371/journal.pone.0200566 URL |
| [10] |
Zhu D, Li RT, Liu X, et al. The positive regulatory roles of the TIFY10 proteins in plant responses to alkaline stress[J]. PLoS One, 2014, 9(11):e111984.
doi: 10.1371/journal.pone.0111984 URL |
| [11] |
Zhao CY, Pan XW, Yu Y, et al. Overexpression of a TIFY family gene, GsJAZ2, exhibits enhanced tolerance to alkaline stress in soybean[J]. Mol Breed, 2020, 40(3):1-13.
doi: 10.1007/s11032-019-1080-6 URL |
| [12] |
Seo JS, Joo J, Kim MJ, et al. OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice[J]. Plant J, 2011, 65(6):907-921.
doi: 10.1111/j.1365-313X.2010.04477.x URL |
| [13] | Yang YX, Ahammed GJ, Wan CP, et al. Comprehensive analysis of TIFY transcription factors and their expression profiles under jasmonic acid and abiotic stresses in watermelon[J]. Int J Genomics, 2019, 2019:6813086. |
| [14] |
Zhang ZB, Li XL, Yu R, et al. Isolation, structural analysis, and expression characteristics of the maize TIFY gene family[J]. Mol Genet Genomics, 2015, 290(5):1849-1858.
doi: 10.1007/s00438-015-1042-6 URL |
| [15] |
Yu YH, Wan YT, Jiao ZL, et al. Functional characterization of resistance to powdery mildew of VvTIFY9 from Vitis vinifera[J]. Int J Mol Sci, 2019, 20(17):4286.
doi: 10.3390/ijms20174286 URL |
| [16] | 刘嘉鹏, 谭秋月, 伍俊为, 等. 香蕉MaTIFY9基因的克隆及表达[J]. 应用与环境生物学报, 2021, 27(6):1609-1615. |
| Liu JP, Tan QY, Wu JW, et al. Cloning and expression analysis of the MaTIFY9 gene in banana[J]. Chin J Appl Environ Biol, 2021, 27(6):1609-1615. | |
| [17] |
胡子曜, 代培红, 刘超, 等. 陆地棉小GTP结合蛋白基因GhROP3的克隆、表达及VIGS载体的构建[J]. 生物技术通报, 2021, 37(9):106-113.
doi: 10.13560/j.cnki.biotech.bull.1985.2021-0287 |
| Hu ZY, Dai PH, Liu C, et al. Molecular cloning, expression and VIGS construction of a small GTP-binding protein gene GhROP3 in Gossypium hirsutum[J]. Biotechnol Bull, 2021, 37(9):106-113. | |
| [18] |
Li Y, Zhou YJ, Dai PH, et al. Cotton Bsr-k1 modulates lignin deposition participating in plant resistance against Verticillium dahliae and Fusarium oxysporum[J]. Plant Growth Regul, 2021, 95(2):283-292.
doi: 10.1007/s10725-021-00742-4 URL |
| [19] |
袁伟, 万红建, 杨悦俭. 植物实时荧光定量PCR内参基因的特点及选择[J]. 植物学报, 2012, 47(4):427-436.
doi: 10.3724/SP.J.1259.2012.00427 |
| Yuan W, Wan HJ, Yang YJ. Characterization and selection of reference genes for real-time quantitative RT-PCR of plants[J]. Chin Bull Bot, 2012, 47(4):427-436. | |
| [20] |
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔct Method[J]. Methods, 2001, 25(4):402-408.
doi: 10.1006/meth.2001.1262 pmid: 11846609 |
| [21] | 王钰静. GhROP6在棉花抗黄萎病中的功能研究[D]. 武汉: 华中农业大学, 2017. |
| Wang YJ. Functional analysis of GhROP6 in cotton responsive to Verticillium dahliae[D]. Wuhan: Huazhong Agricultural University, 2017. | |
| [22] |
Zhang DD, Wang J, Wang D, et al. Population genomics demystifies the defoliation phenotype in the plant pathogen Verticillium dahliae[J]. New Phytol, 2019, 222(2):1012-1029.
doi: 10.1111/nph.15672 URL |
| [23] |
Hrmova M, Hussain SS. Plant transcription factors involved in drought and associated stresses[J]. Int J Mol Sci, 2021, 22(11):5662.
doi: 10.3390/ijms22115662 URL |
| [24] |
Yu FF, Huaxia YF, Lu WJ, et al. GhWRKY15, a member of the WR-KY transcription factor family identified from cotton(Gossypium hirsutum L. ), is involved in disease resistance and plant develop-ment[J]. BMC Plant Biol, 2012, 12:144.
doi: 10.1186/1471-2229-12-144 URL |
| [25] |
Ma Q, Wang NH, Ma L, et al. The cotton BEL1-like transcription factor GhBLH7-D06 negatively regulates the defense response against Verticillium dahliae[J]. Int J Mol Sci, 2020, 21(19):7126.
doi: 10.3390/ijms21197126 URL |
| [26] |
Hu Q, Xiao SH, Wang XR, et al. GhWRKY1-like enhances cotton resistance to Verticillium dahliae via an increase in defense-induced lignification and S monolignol content[J]. Plant Sci, 2021, 305:110833.
doi: 10.1016/j.plantsci.2021.110833 URL |
| [27] |
Xiao SH, Hu Q, Shen JL, et al. GhMYB4 downregulates lignin biosynthesis and enhances cotton resistance to Verticillium dahliae[J]. Plant Cell Rep, 2021, 40(4):735-751.
doi: 10.1007/s00299-021-02672-x URL |
| [28] |
He X, Wang TY, Zhu W, et al. GhHB12, a HD-ZIP I transcription factor, negatively regulates the cotton resistance to Verticillium dahliae[J]. Int J Mol Sci, 2018, 19(12):3997.
doi: 10.3390/ijms19123997 URL |
| [29] | Liu TL, Chen TZ, Kan JL, et al. The GhMYB36 transcription factor confers resistance to biotic and abiotic stress by enhancing PR1 gene expression in plants[J]. Plant Biotechnol J, 2021:2021 Nov 23. |
| [30] |
Xiong XP, Sun SC, Zhang XY, et al. GhWRKY70D13 regulates resistance to Verticillium dahliae in cotton through the ethylene and jasmonic acid signaling pathways[J]. Front Plant Sci, 2020, 11:69.
doi: 10.3389/fpls.2020.00069 URL |
| [31] |
He X, Zhu LF, Xu L, et al. GhATAF1, a NAC transcription factor, confers abiotic and biotic stress responses by regulating phytohormonal signaling networks[J]. Plant Cell Rep, 2016, 35(10):2167-2179.
doi: 10.1007/s00299-016-2027-6 URL |
| [32] |
Cheng HQ, Han LB, Yang CL, et al. The cotton MYB108 forms a positive feedback regulation loop with CML11 and participates in the defense response against Verticillium dahliae infection[J]. J Exp Bot, 2016, 67(6):1935-1950.
doi: 10.1093/jxb/erw016 URL |
| [33] |
Zhang K, Zhao P, Wang HM, et al. Isolation and characterization of the GbVIP1 gene and response to Verticillium wilt in cotton and tobacco[J]. J Cotton Res, 2019, 2:2.
doi: 10.1186/s42397-019-0019-0 URL |
| [34] |
Guo WF, Jin L, Miao YH, et al. An ethylene response-related factor, GbERF1-like, from Gossypium barbadense improves resistance to Verticillium dahliae via activating lignin synthesis[J]. Plant Mol Biol, 2016, 91(3):305-318.
doi: 10.1007/s11103-016-0467-6 URL |
| [35] |
Li XC, Liu NN, Sun Y, et al. The cotton GhWIN2 gene activates the cuticle biosynthesis pathway and influences the salicylic and jasmonic acid biosynthesis pathways[J]. BMC Plant Biol, 2019, 19(1):379.
doi: 10.1186/s12870-019-1888-6 URL |
| [36] |
He X, Zhu LF, Wassan GM, et al. GhJAZ2 attenuates cotton resistance to biotic stresses via the inhibition of the transcriptional activity of GhbHLH171[J]. Mol Plant Pathol, 2018, 19(4):896-908.
doi: 10.1111/mpp.12575 URL |
| [37] | 张祥瑞. 棉花抗黄萎病相关基因GhJAZ4和GhbHLH25的功能初探[D]. 开封: 河南大学, 2018. |
| Zhang XR. Preliminary study on functions of GhJAZ4 and GhbHLH25 genes related to Verticillium wilt resistance in cotton[D]. Kaifeng: Henan University, 2018. | |
| [38] |
Song Y, Zhai YH, Li LX, et al. BIN2 negatively regulates plant defence against Verticillium dahliae in Arabidopsis and cotton[J]. Plant Biotechnol J, 2021, 19(10):2097-2112.
doi: 10.1111/pbi.13640 URL |
| [1] | 吕秋谕, 孙培媛, 冉彬, 王佳蕊, 陈庆富, 李洪有. 苦荞转录因子基因FtbHLH3的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 194-203. |
| [2] | 王佳蕊, 孙培媛, 柯瑾, 冉彬, 李洪有. 苦荞糖基转移酶基因FtUGT143的克隆及表达分析[J]. 生物技术通报, 2023, 39(8): 204-212. |
| [3] | 孙明慧, 吴琼, 刘丹丹, 焦小雨, 王文杰. 茶树CsTMFs的克隆与表达分析[J]. 生物技术通报, 2023, 39(7): 151-159. |
| [4] | 刘思佳, 王浩楠, 付宇辰, 闫文欣, 胡增辉, 冷平生. ‘西伯利亚’百合LiCMK基因克隆及功能分析[J]. 生物技术通报, 2023, 39(3): 196-205. |
| [5] | 杨旭妍, 赵爽, 马天意, 白玉, 王玉书. 三个甘蓝WRKY基因的克隆及其对非生物胁迫的表达[J]. 生物技术通报, 2023, 39(11): 261-269. |
| [6] | 侯瑞泽, 鲍悦, 陈启亮, 毛桂玲, 韦博霖, 侯雷平, 李梅兰. 普通白菜PRR5的克隆、表达及功能验证[J]. 生物技术通报, 2023, 39(10): 128-135. |
| [7] | 杨敏, 龙雨青, 曾娟, 曾梅, 周新茹, 王玲, 付学森, 周日宝, 刘湘丹. 灰毡毛忍冬UGTPg17、UGTPg36基因克隆及功能研究[J]. 生物技术通报, 2023, 39(10): 256-267. |
| [8] | 王楠, 张瑞, 潘阳阳, 何翃宏, 王靖雷, 崔燕, 余四九. 牦牛TGF-β1基因克隆及在雌性生殖系统主要器官中的表达定位[J]. 生物技术通报, 2022, 38(6): 279-290. |
| [9] | 李洋, 张晓天, 朴静子, 周如军, 李自博, 关海雯. 花生疮痂病菌蓝光受体EaWC 1基因克隆及生物信息学分析[J]. 生物技术通报, 2022, 38(5): 93-99. |
| [10] | 胡琪, 侯玉翔, 李璿, 李梅兰. 普通白菜CYP79B2同源基因的克隆与表达[J]. 生物技术通报, 2022, 38(12): 168-174. |
| [11] | 甘诚燕, 张心慧, 王沙, 樊瑶羽薇, 招雪晴, 苑兆和. 石榴花发育相关基因PgSPL2的克隆及功能研究[J]. 生物技术通报, 2022, 38(12): 194-203. |
| [12] | 盛雪晴, 赵楠, 林亚秋, 陈定双, 王瑞龙, 李傲, 王永, 李艳艳. 山羊ZNF32的克隆及表达分析[J]. 生物技术通报, 2022, 38(12): 300-311. |
| [13] | 付伟杰, 邝杰华, 罗君, 黄建盛, 陈有铭, 陈刚. 杉虎斑Galectin-8基因克隆及其在不同阿魏酸水平饲料下的表达响应[J]. 生物技术通报, 2022, 38(12): 312-323. |
| [14] | 张琳, 魏祯祯, 宋程威, 郭丽丽, 郭琪, 侯小改, 王华芳. ‘凤丹’牡丹PoFD基因克隆及表达分析[J]. 生物技术通报, 2022, 38(11): 104-111. |
| [15] | 党瑗, 李维, 苗向, 修宇, 林善枝. 山杏油体蛋白基因PsOLE4克隆及其调控油脂累积功能分析[J]. 生物技术通报, 2022, 38(11): 151-161. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||