








生物技术通报 ›› 2022, Vol. 38 ›› Issue (8): 118-126.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1192
收稿日期:2021-09-15
出版日期:2022-08-26
发布日期:2022-09-14
作者简介:黄婧,女,博士,助理研究员,研究方向:植物重金属修复、植物生长发育;E-mail: 基金资助:
HUANG Jing1( ), ZHU Liang1,2, XUE Peng-bo1,2, FU Qiang1(
), ZHU Liang1,2, XUE Peng-bo1,2, FU Qiang1( )
)
Received:2021-09-15
Published:2022-08-26
Online:2022-09-14
摘要:
水稻是我国最重要的粮食作物,挖掘控制水稻叶和籽粒镉积累的关键基因,阐明水稻镉积累和转运的遗传机制,为培育籽粒低镉积累的水稻品种、保障粮食安全和人类健康提供依据。通过离子组学技术系统分析中国栽培稻核心种质资源209个品种籽粒的离子谱,鉴定出一批铁、锌等高富集以及镉低积累的水稻品种,并选择籼稻(indica)品种花楸03和粳稻(japonica)品种SKC进行进一步研究。结果表明,尽管这两个品种对镉的耐受性和吸收能力无显著差异,花楸03籽粒和地上部分的镉积累量显著高于SKC,而铁、锌等含量差异不显著。以花楸03和SKC为亲本构建了包含137个单株的双单倍体(doubled haploid,DH)群体,共检测到8个控制镉在叶和籽粒中积累的QTL,分别位于第2、3、4、7、8、10和11染色体上,能解释10.6%-39.4%的表型变异。其中第3染色体上检测到的控制镉向籽粒转运的QTL qGCd3定位在RM6266-RM2334,LOD(limit of detection)值和贡献率分别为3.81和39.4%,此处可能存在一个控制镉向籽粒转运的重要基因。
黄婧, 朱亮, 薛蓬勃, 付强. 水稻叶和籽粒镉积累机制及QTL定位研究[J]. 生物技术通报, 2022, 38(8): 118-126.
HUANG Jing, ZHU Liang, XUE Peng-bo, FU Qiang. Research on Mechanism and QTL Mapping Associated with Cadmium Accumulation in Rice Leaves and Grains[J]. Biotechnology Bulletin, 2022, 38(8): 118-126.
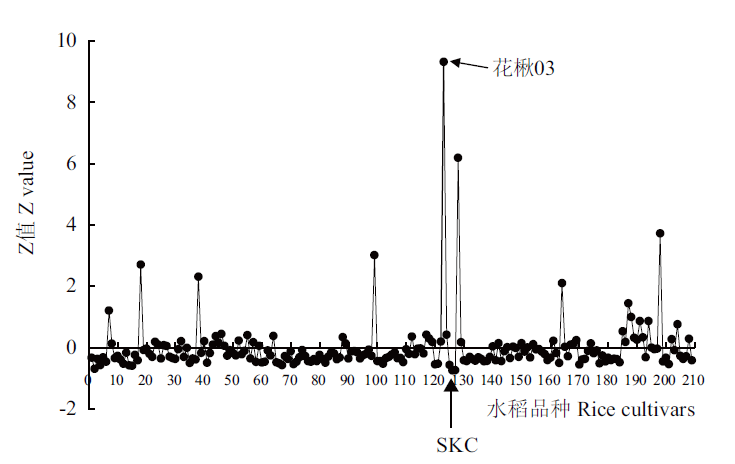
图1 核心种质资源水稻籽粒中Cd的积累 Z值=(样本元素浓度-平均值)/标准差,用于表达样品中某一元素偏离群体平均值的程度。X轴表示水稻品种的编号
Fig. 1 Profiling of Cd accumulation in grains of rice core germplasm Z value =(individual value-population average)/population SD,and represents SD from the average value of the whole population. X axis refers to the serial number of rice cultivars
| 土壤类型Soil type | 镉Cd | 铁Fe | 锌Zn | 铜Cu | 锰Mn | 砷As |
|---|---|---|---|---|---|---|
| 对照水槽Control sink | 0.14±0.02 | 19 047.64±232.07 | 157.34±8.26 | 23.51±9.55 | 318.16±3.63 | 9.69±2.69 |
| 污染水槽 Contaminated sink | 1.09±0.11 | 20 495.83±270.84 | 271.98±9.51 | 45.39±8.31 | 322.02±9.58 | 12.99±1.28 |
| 污染大田土壤Contaminated paddy field | 0.55±0.07 | 19 603.91±267.35 | 265.89±4.15 | 48.20±3.93 | 330.8±14.11 | 12.53±1.34 |
表1 大田、镉污染水槽和对照水槽土壤中Cd等元素的含量
Table 1 Contents of Cd and other metals in the soil of contaminated paddy field,contaminated sink and control sink (μg·g-1 DW)
| 土壤类型Soil type | 镉Cd | 铁Fe | 锌Zn | 铜Cu | 锰Mn | 砷As |
|---|---|---|---|---|---|---|
| 对照水槽Control sink | 0.14±0.02 | 19 047.64±232.07 | 157.34±8.26 | 23.51±9.55 | 318.16±3.63 | 9.69±2.69 |
| 污染水槽 Contaminated sink | 1.09±0.11 | 20 495.83±270.84 | 271.98±9.51 | 45.39±8.31 | 322.02±9.58 | 12.99±1.28 |
| 污染大田土壤Contaminated paddy field | 0.55±0.07 | 19 603.91±267.35 | 265.89±4.15 | 48.20±3.93 | 330.8±14.11 | 12.53±1.34 |
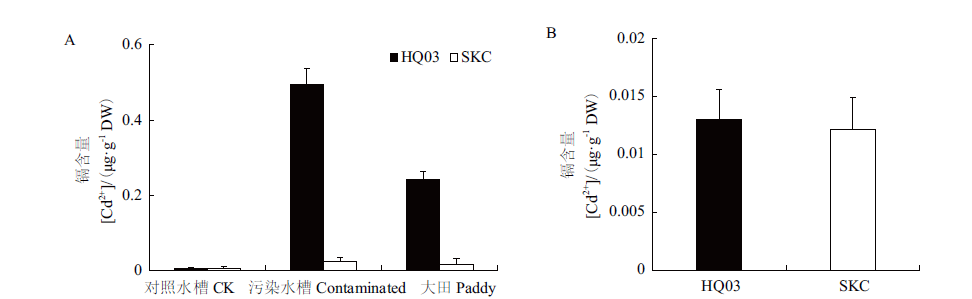
图3 花楸03与SKC的籽粒和精米中的Cd含量 A:分别种植在污染大田、污染水槽和对照水槽中,花楸03与SKC籽粒中的Cd含量;B:种植在镉污染水槽中,花楸03与SKC精米中的Cd含量。**:P<0.01。下同
Fig.3 Cd accumulation in the grains and milled rice of HQ03 and SKC A:Cd accumulation in the grains of HQ03 and SKC from the rice grown in the contaminated paddy field,contaminated and control sink,respectively. B:Cd accumulation in the milled rice of HQ03 and SKC from the rice grown in the contaminated sink. **:P < 0.01. The same below
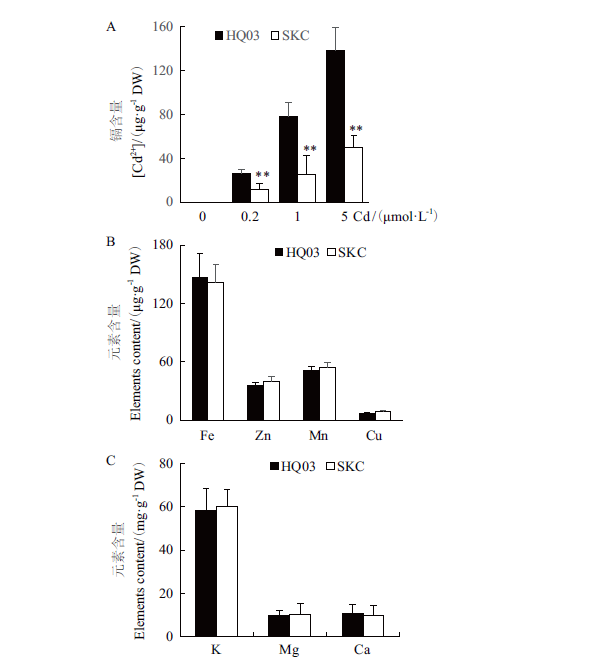
图4 花楸03与SKC叶片中的元素积累 A:花楸03和SKC叶中的Cd含量;B、C:花楸03和SKC叶中的Fe等金属元素的含量(B)和K等大量元素的含量(C)
Fig.4 Elements accumulation in the leaves of HQ03 and SKC A:Cd accumulation in the leaves of HQ03 and SKC. B, C:Contents of metal elements such Fe(B)and major elements such as K(C)in the leaves of HQ03 and SKC
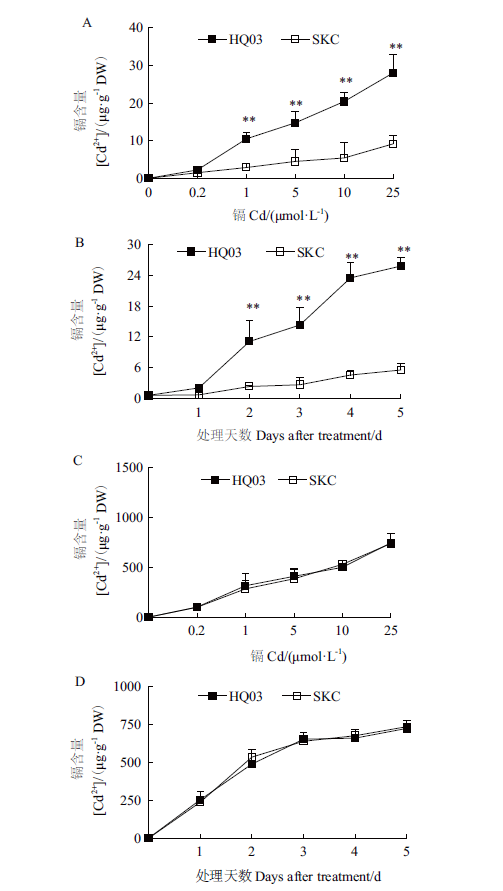
图5 不同浓度和不同时间CdCl2处理后花楸03和SKC幼苗叶片与根中Cd的积累 A:不同浓度CdCl2处理7 d后,叶片中Cd的含量;B:用10 μmol/L CdCl2处理不同时间后,叶片中Cd的含量;C:不同浓度CdCl2处理7 d后,根中Cd的含量;D:用10 μmol/L CdCl2处理不同时间后,根中Cd的含量
Fig.5 Cd accumulation in the leaves and roots of HQ03 and SKC from rice seedlings exposed to CdCl2 for indicated concentrations and days A:Cd levels in leaves from rice seedlings exposed to CdCl2 for 7 d at indicated concentrations;B:Cd levels in leaves from rice seedlings exposed to 10 μmol/L CdCl2 for indicated days;C:Cd levels in roots from rice seedlings exposed to CdCl2 for 7 d at indicated concentrations;D:Cd levels in roots from rice seedlings exposed to 10 μmol/L CdCl2 for indicated days
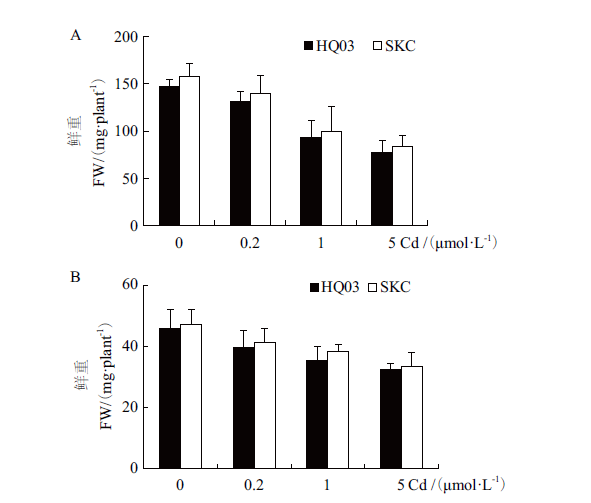
图6 花楸03与SKC对Cd的耐受性分析 A:不同浓度Cd处理下,花楸03和SKC地上部鲜重(FW);B:不同浓度Cd处理下,花楸03和SKC地下部鲜重(FW)
Fig. 6 Tolerance assay of HQ03 and SKC to Cd A:Fresh weight(FW)of shoots of HQ03 and SKC under different concentrations of Cd treatment. B:Fresh weight(FW)of roots of HQ03 and SKC under different concentrations of Cd treatment
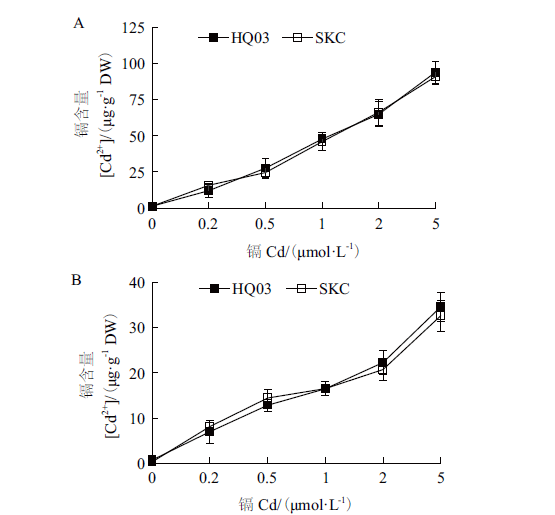
图7 花楸03和SKC的根吸收试验 A:正常培养条件(28℃);B:低温条件(4℃)
Fig.7 Root uptake assay of HQ03 and SKC A:Normal culture condition(28℃). B:Low temperature culture condition(4℃)
| 数量性状基因座位QTL | 染色体Chr. | 标记区间Marker interval | LOD值LOD value | 贡献率 R2/% | 加性效应Additive effect |
|---|---|---|---|---|---|
| qLCd2 | 2 | RM450-RM5472 | 2.70 | 13.4 | 12.792 |
| qLCd4 | 4 | RM307-RM401 | 2.14 | 11.1 | 11.234 |
| qLCd10 | 10 | RM271-RM258 | 3.02 | 10.6 | -0.081 8 |
| qLCd11 | 11 | RM167-RM220 | 2.07 | 11.0 | 7.576 1 |
表2 水稻叶Cd积累的QTL定位
Table 2 Quantitative trait loci(QTLs)mapping for Cd accumulation in rice leaves
| 数量性状基因座位QTL | 染色体Chr. | 标记区间Marker interval | LOD值LOD value | 贡献率 R2/% | 加性效应Additive effect |
|---|---|---|---|---|---|
| qLCd2 | 2 | RM450-RM5472 | 2.70 | 13.4 | 12.792 |
| qLCd4 | 4 | RM307-RM401 | 2.14 | 11.1 | 11.234 |
| qLCd10 | 10 | RM271-RM258 | 3.02 | 10.6 | -0.081 8 |
| qLCd11 | 11 | RM167-RM220 | 2.07 | 11.0 | 7.576 1 |
| 数量性状基因座位QTL | 染色体Chr. | 标记区间Marker interval | LOD值LOD value | 贡献率R2/% | 加性效应Additive effect |
|---|---|---|---|---|---|
| qGCd2 | 2 | RM324-RM341 | 3.39 | 20.2 | -0.187 4 |
| qGCd3 | 3 | RM6266-RM2334 | 3.81 | 39.4 | 0.125 4 |
| qGCd7 | 7 | RM6574-RM6449 | 2.51 | 13.2 | -5.381 7 |
| qGCd8 | 8 | RM1235-RM1376 | 3.15 | 20.4 | -0.278 1 |
表3 水稻籽粒Cd积累的QTL定位
Table 3 Quantitative trait loci(QTLs)mapping for Cd accumulation in rice grains
| 数量性状基因座位QTL | 染色体Chr. | 标记区间Marker interval | LOD值LOD value | 贡献率R2/% | 加性效应Additive effect |
|---|---|---|---|---|---|
| qGCd2 | 2 | RM324-RM341 | 3.39 | 20.2 | -0.187 4 |
| qGCd3 | 3 | RM6266-RM2334 | 3.81 | 39.4 | 0.125 4 |
| qGCd7 | 7 | RM6574-RM6449 | 2.51 | 13.2 | -5.381 7 |
| qGCd8 | 8 | RM1235-RM1376 | 3.15 | 20.4 | -0.278 1 |
| [1] |
Haider FU, Liqun C, Coulter JA, et al. Cadmium toxicity in plants:Impacts and remediation strategies[J]. Ecotoxicol Environ Saf, 2021, 211:111887.
doi: 10.1016/j.ecoenv.2020.111887 URL |
| [2] | Chen HM, Chen WC, Chen YX, et al. Pollution of heavy metals in soil-plant system[M]. Beijing: Science Press, 1996. |
| [3] |
Song Y, Wang Y, Mao WF, et al. Dietary cadmium exposure assessment among the Chinese population[J]. PLoS One, 2017, 12(5):e0177978.
doi: 10.1371/journal.pone.0177978 URL |
| [4] |
Nawrot TS, Staessen JA, Roels HA, et al. Cadmium exposure in the population:from health risks to strategies of prevention[J]. Biometals, 2010, 23(5):769-782.
doi: 10.1007/s10534-010-9343-z pmid: 20517707 |
| [5] |
Meng L, Huang TH, Shi JC, et al. Decreasing cadmium uptake of rice(Oryza sativa L.)in the cadmium-contaminated paddy field through different cultivars coupling with appropriate soil amendments[J]. J Soils Sediments, 2019, 19(4):1788-1798.
doi: 10.1007/s11368-018-2186-x URL |
| [6] |
Mendoza-Cózatl DG, Jobe TO, Hauser F, et al. Long-distance transport, vacuolar sequestration, tolerance, and transcriptional responses induced by cadmium and arsenic[J]. Curr Opin Plant Biol, 2011, 14(5):554-562.
doi: 10.1016/j.pbi.2011.07.004 pmid: 21820943 |
| [7] | Zhao JL, Yang W, Zhang SH, et al. Genome-wide association study and candidate gene analysis of rice cadmium accumulation in grain in a diverse rice collection[J]. Rice:N Y, 2018, 11(1):61. |
| [8] |
Xue DW, Chen MC, Zhang GP. Mapping of QTLs associated with cadmium tolerance and accumulation during seedling stage in rice(Oryza sativa L.)[J]. Euphytica, 2008, 165(3):587-596.
doi: 10.1007/s10681-008-9785-3 URL |
| [9] |
Ueno D, Kono I, Yokosho K, et al. A major quantitative trait locus controlling cadmium translocation in rice(Oryza sativa)[J]. New Phytol, 2009, 182(3):644-653.
doi: 10.1111/j.1469-8137.2009.02784.x pmid: 19309445 |
| [10] |
Ishikawa S, Abe T, Kuramata M, et al. A major quantitative trait locus for increasing cadmium-specific concentration in rice grain is located on the short arm of chromosome 7[J]. J Exp Bot, 2010, 61(3):923-934.
doi: 10.1093/jxb/erp360 pmid: 20022924 |
| [11] |
Ueno D, Yamaji N, Kono I, et al. Gene limiting cadmium accumulation in rice[J]. Proc Natl Acad Sci USA, 2010, 107(38):16500-16505.
doi: 10.1073/pnas.1005396107 URL |
| [12] |
Miyadate H, Adachi S, Hiraizumi A, et al. OsHMA3, a P1B-type of ATPase affects root-to-shoot cadmium translocation in rice by mediating efflux into vacuoles[J]. New Phytol, 2011, 189(1):190-199.
doi: 10.1111/j.1469-8137.2010.03459.x URL |
| [13] |
Huang Y, Sun CX, Min J, et al. Association mapping of quantitative trait loci for mineral element contents in whole grain rice(Oryza sativa L.)[J]. J Agric Food Chem, 2015, 63(50):10885-10892.
doi: 10.1021/acs.jafc.5b04932 URL |
| [14] |
Wang FJ, Wang M, Liu ZB, et al. Different responses of low grain-Cd-accumulating and high grain-Cd-accumulating rice cultivars to Cd stress[J]. Plant Physiol Biochem, 2015, 96:261-269.
doi: 10.1016/j.plaphy.2015.08.001 URL |
| [15] |
Luo JS, Huang J, Zeng DL, et al. A defensin-like protein drives cadmium efflux and allocation in rice[J]. Nat Commun, 2018, 9(1):645.
doi: 10.1038/s41467-018-03088-0 URL |
| [16] |
Hu DW, Sheng ZH, Li QL, et al. Identification of QTLs associated with cadmium concentration in rice grains[J]. J Integr Agric, 2018, 17(7):1563-1573.
doi: 10.1016/S2095-3119(17)61847-1 URL |
| [17] |
Liu WQ, Pan XW, Li YC, et al. Identification of QTLs and validation of qCd-2 associated with grain cadmium concentrations in rice[J]. Rice Sci, 2019, 26(1):42-49.
doi: 10.1016/j.rsci.2018.12.003 URL |
| [18] |
Yan HL, Xu WX, Xie JY, et al. Variation of a major facilitator superfamily gene contributes to differential cadmium accumulation between rice subspecies[J]. Nat Commun, 2019, 10(1):2562.
doi: 10.1038/s41467-019-10544-y URL |
| [19] |
Pan XW, Li YC, Liu WQ, et al. QTL mapping and candidate gene analysis of cadmium accumulation in polished rice by genome-wide association study[J]. Sci Rep, 2020, 10(1):11791.
doi: 10.1038/s41598-020-68742-4 URL |
| [20] | Lincoln SE, Daly MJ, Lander ES. Constructing genetic linkage maps with mapmaker/Exp version 3.0: A tutorial and reference manual[M]. 3rd ed. Cambridge: Whitehead Institute for Biometrical Research, 1993. |
| [21] | Lincoln SE, Daly MJ, Lander ES. Mapping genes controlling quantitative traits using MAPMAKER/QTL version 1.1: A tutorial and reference manual[M]. 2nd ed. Cambridge: Whitehead Institute for Biometrical Research, 1993. |
| [22] |
Uraguchi S, Mori S, Kuramata M, et al. Root-to-shoot Cd translocation via the xylem is the major process determining shoot and grain cadmium accumulation in rice[J]. J Exp Bot, 2009, 60(9):2677-2688.
doi: 10.1093/jxb/erp119 pmid: 19401409 |
| [23] |
Takahashi A, Agrawal GK, Yamazaki M, et al. Rice Pti1a negatively regulates RAR1-dependent defense responses[J]. Plant Cell, 2007, 19(9):2940-2951.
pmid: 17890377 |
| [1] | 王子颖, 龙晨洁, 范兆宇, 张蕾. 利用酵母双杂交系统筛选水稻中与OsCRK5互作蛋白[J]. 生物技术通报, 2023, 39(9): 117-125. |
| [2] | 黄小龙, 孙贵连, 马丹丹, 闫慧清. 水稻幼苗酵母单杂文库构建及LAZY1上游调控因子筛选[J]. 生物技术通报, 2023, 39(9): 126-135. |
| [3] | 李雪琪, 张素杰, 于曼, 黄金光, 周焕斌. 基于CRISPR/CasX介导的水稻基因组编辑技术的建立[J]. 生物技术通报, 2023, 39(9): 40-48. |
| [4] | 吴元明, 林佳怡, 柳雨汐, 李丹婷, 张宗琼, 郑晓明, 逄洪波. 基于BSA-seq和RNA-seq挖掘水稻株高相关QTL[J]. 生物技术通报, 2023, 39(8): 173-184. |
| [5] | 姚莎莎, 王晶晶, 王俊杰, 梁卫红. 植物激素信号通路调控水稻粒型的分子机制[J]. 生物技术通报, 2023, 39(8): 80-90. |
| [6] | 徐建霞, 丁延庆, 冯周, 曹宁, 程斌, 高旭, 邹桂花, 张立异. 基于Super-GBS的高粱株高和节间数QTL定位[J]. 生物技术通报, 2023, 39(7): 185-194. |
| [7] | 余慧, 王静, 梁昕昕, 辛亚平, 周军, 赵会君. 宁夏枸杞铁镉响应基因的筛选及其功能验证[J]. 生物技术通报, 2023, 39(7): 195-205. |
| [8] | 李宇, 李素贞, 陈茹梅, 卢海强. 植物bHLH转录因子调控铁稳态的研究进展[J]. 生物技术通报, 2023, 39(7): 26-36. |
| [9] | 刘辉, 卢扬, 叶夕苗, 周帅, 李俊, 唐健波, 陈恩发. 外源硫诱导苦荞镉胁迫响应的比较转录组学分析[J]. 生物技术通报, 2023, 39(5): 177-191. |
| [10] | 任沛东, 彭健玲, 刘圣航, 姚姿婷, 朱桂宁, 陆光涛, 李瑞芳. 沙福芽孢杆菌GX-H6的分离鉴定及对水稻细菌性条斑病的防病效果[J]. 生物技术通报, 2023, 39(5): 243-253. |
| [11] | 李怡君, 吴晨晨, 李睿, 王喆, 何山文, 韦善君, 张晓霞. 水稻内生细菌新资源分离培养方案探究[J]. 生物技术通报, 2023, 39(4): 201-211. |
| [12] | 卢振万, 李雪琪, 黄金光, 周焕斌. 利用胞嘧啶碱基编辑技术创制耐草甘膦水稻[J]. 生物技术通报, 2023, 39(2): 63-69. |
| [13] | 杨茂, 林宇丰, 戴阳朔, 潘素君, 彭伟业, 严明雄, 李魏, 王冰, 戴良英. OsDIS1通过抗氧化途径负调控水稻耐旱性[J]. 生物技术通报, 2023, 39(2): 88-95. |
| [14] | 蒋铭轩, 李康, 罗亮, 刘建祥, 芦海平. 植物表达外源蛋白研究进展及展望[J]. 生物技术通报, 2023, 39(11): 110-122. |
| [15] | 姜南, 石杨, 赵志慧, 李斌, 赵熠辉, 杨俊彪, 闫家铭, 靳雨璠, 陈稷, 黄进. 镉胁迫下水稻OsPT1的表达及功能分析[J]. 生物技术通报, 2023, 39(1): 166-174. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||