








生物技术通报 ›› 2023, Vol. 39 ›› Issue (2): 263-273.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0689
曲春娟1( ), 朱悦1,2, 江晨1, 曲明静1, 王向誉3(
), 朱悦1,2, 江晨1, 曲明静1, 王向誉3( ), 李晓1(
), 李晓1( )
)
收稿日期:2022-06-04
出版日期:2023-02-26
发布日期:2023-03-07
作者简介:曲春娟,女,助理研究员,研究方向:农业昆虫学;E-mail: 基金资助:
QU Chun-juan1( ), ZHU Yue1,2, JIANG Chen1, QU Ming-jing1, WANG Xiang-yu3(
), ZHU Yue1,2, JIANG Chen1, QU Ming-jing1, WANG Xiang-yu3( ), LI Xiao1(
), LI Xiao1( )
)
Received:2022-06-04
Published:2023-02-26
Online:2023-03-07
摘要:
铜绿丽金龟Anomala corpulenta是一种重要的农林害虫。本文采用Illumina HiSeq X平台对其进行线粒体基因组测序,对基因组序列进行拼装、注释,并对线粒体基因组结构特征和碱基组成进行分析。基于13个蛋白质编码基因的核苷酸序列,采用最大似然法和贝叶斯法构建金龟科系统发育树。结果表明,铜绿丽金龟线粒体基因组全长为16 673 bp,包括13个蛋白质编码基因、22个tRNA基因、2个rRNA基因和一段长约1 074 bp的A+T富集区。基因排布与已知的其他丽金龟科昆虫完全相同,且遵循祖先模式,未发生基因重排。系统发育分析显示,丽金龟亚科的所有物种聚在同一分支,支持该亚科的单系性;异丽金龟属Anomala与彩丽金龟属Mimela的亲缘关系较其与弧丽金龟Popillia和喙丽金龟属Adoretus更近。本研究获得了第一条丽金龟亚科最大属——异丽金龟属Anomala昆虫的线粒体基因组全序列,有助于加深对丽金龟亚科线粒体基因组学和系统发育关系的理解。
曲春娟, 朱悦, 江晨, 曲明静, 王向誉, 李晓. 铜绿丽金龟线粒体全基因组及其系统发育分析[J]. 生物技术通报, 2023, 39(2): 263-273.
QU Chun-juan, ZHU Yue, JIANG Chen, QU Ming-jing, WANG Xiang-yu, LI Xiao. Whole Mitochondrial Genome and Phylogeny Analysis of Anomala corpulenta[J]. Biotechnology Bulletin, 2023, 39(2): 263-273.
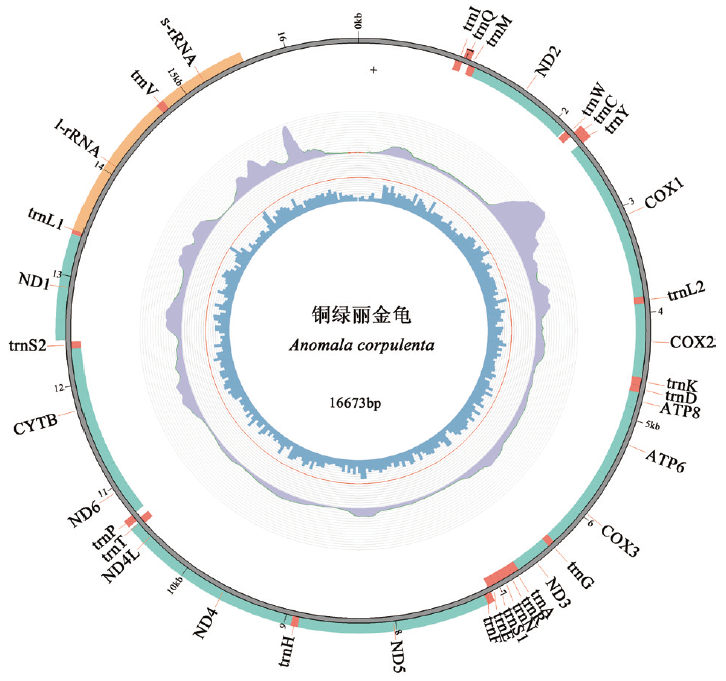
图1 铜绿丽金龟线粒体基因组结构图 从外到内依次为:基因组结构图,reads在基因组上的覆盖度,GC含量
Fig. 1 Mitochondrial genome structure of A. corpulenta From outside to inside: Genome structure, coverage of reads on the genome, and GC content
| 基因 Gene | 位置 Position/bp | 编码链 Coding strand | 长度 Length/bp | 起始密码子 Start codon | 终止密码子 Stop codon | 反密码子 Anticodon | 基因间隔 Intergenic length/bp |
|---|---|---|---|---|---|---|---|
| tRNAIle | 915-979 | J | 65 | GAT | - | ||
| tRNAGln | 976-1 045 | N | 70 | TTG | -4 | ||
| tRNAMet | 1 044-1 113 | J | 70 | CAT | -2 | ||
| nad2 | 1 113-2 121 | J | 1 009 | ATG | TAA | -1 | |
| tRNATrp | 2 135-2 203 | J | 69 | TCA | 13 | ||
| tRNACys | 2 195-2 257 | N | 63 | GCA | -9 | ||
| tRNATyr | 2 257-2 322 | N | 66 | GTA | -1 | ||
| cox1 | 2 314-3 859 | J | 1 546 | ATT | TAA | -9 | |
| tRNALeu(UUR) | 3 854-3 919 | J | 66 | TAA | -6 | ||
| cox2 | 3 919-4 627 | J | 709 | ATC | TAA | -1 | |
| tRNALys | 4 607-4 678 | J | 72 | CTT | -21 | ||
| tRNAAsp | 4 680-4 745 | J | 66 | GTC | 1 | ||
| atp8 | 4 745-4 901 | J | 157 | ATT | TAA | -1 | |
| atp6 | 4 897-5 569 | J | 673 | ATA | TAA | -5 | |
| cox3 | 5 568-6 356 | J | 789 | ATG | TA(A) | -2 | |
| tRNAGly | 6 355-6 420 | J | 66 | TCC | -2 | ||
| nad3 | 6 420-6 774 | J | 355 | ATC | TAG | -1 | |
| tRNAAla | 6 772-6 836 | J | 65 | TGC | -2 | ||
| tRNAArg | 6 836-6 901 | J | 66 | TCG | -1 | ||
| tRNAAsn | 6 901-6 966 | J | 66 | GTT | -1 | ||
| tRNASer(AGN) | 6 966-7 033 | J | 68 | TCT | -1 | ||
| tRNAGlu | 7 033-7 098 | J | 66 | TTC | -1 | ||
| tRNAPhe | 7 096-7 163 | N | 68 | GAA | -3 | ||
| nad5 | 7 162-8 878 | N | 1 717 | ATT | TAA | -2 | |
| tRNAHis | 8 881-8 944 | N | 64 | GTG | 2 | ||
| nad4 | 8 943-10 280 | N | 1 338 | ATG | TA(A) | -2 | |
| nad4l | 10 273-10 606 | N | 334 | TTG | TAA | -8 | |
| tRNAThr | 10 566-10 631 | J | 66 | TGT | -41 | ||
| tRNAPro | 10 631-10 696 | N | 66 | TGG | -1 | ||
| nad6 | 10 697-11 201 | J | 505 | ATC | TAA | 0 | |
| cytb | 11 200-12 343 | J | 1 144 | ATG | TAG | -2 | |
| tRNASer(UCN) | 12 341-12 406 | J | 66 | TGA | -3 | ||
| nad1 | 12 422-13 370 | N | 949 | ATA | TAG | 15 | |
| tRNALeu(CUN) | 13 374-13 440 | N | 67 | TAG | 3 | ||
| 16S rRNA | 13 404-14 753 | N | 1 350 | -37 | |||
| tRNAVal | 14 731-14 801 | N | 71 | TAC | -23 | ||
| 12S rRNA | 14 801-15 599 | N | 800 | -1 | |||
| Control Region | 15 600-16 673 | J | 1 074 |
表1 铜绿丽金龟线粒体基因位置与起始终止密码子
Table 1 Locations and start/stop codons of mitochondrial genes in A. corpulenta
| 基因 Gene | 位置 Position/bp | 编码链 Coding strand | 长度 Length/bp | 起始密码子 Start codon | 终止密码子 Stop codon | 反密码子 Anticodon | 基因间隔 Intergenic length/bp |
|---|---|---|---|---|---|---|---|
| tRNAIle | 915-979 | J | 65 | GAT | - | ||
| tRNAGln | 976-1 045 | N | 70 | TTG | -4 | ||
| tRNAMet | 1 044-1 113 | J | 70 | CAT | -2 | ||
| nad2 | 1 113-2 121 | J | 1 009 | ATG | TAA | -1 | |
| tRNATrp | 2 135-2 203 | J | 69 | TCA | 13 | ||
| tRNACys | 2 195-2 257 | N | 63 | GCA | -9 | ||
| tRNATyr | 2 257-2 322 | N | 66 | GTA | -1 | ||
| cox1 | 2 314-3 859 | J | 1 546 | ATT | TAA | -9 | |
| tRNALeu(UUR) | 3 854-3 919 | J | 66 | TAA | -6 | ||
| cox2 | 3 919-4 627 | J | 709 | ATC | TAA | -1 | |
| tRNALys | 4 607-4 678 | J | 72 | CTT | -21 | ||
| tRNAAsp | 4 680-4 745 | J | 66 | GTC | 1 | ||
| atp8 | 4 745-4 901 | J | 157 | ATT | TAA | -1 | |
| atp6 | 4 897-5 569 | J | 673 | ATA | TAA | -5 | |
| cox3 | 5 568-6 356 | J | 789 | ATG | TA(A) | -2 | |
| tRNAGly | 6 355-6 420 | J | 66 | TCC | -2 | ||
| nad3 | 6 420-6 774 | J | 355 | ATC | TAG | -1 | |
| tRNAAla | 6 772-6 836 | J | 65 | TGC | -2 | ||
| tRNAArg | 6 836-6 901 | J | 66 | TCG | -1 | ||
| tRNAAsn | 6 901-6 966 | J | 66 | GTT | -1 | ||
| tRNASer(AGN) | 6 966-7 033 | J | 68 | TCT | -1 | ||
| tRNAGlu | 7 033-7 098 | J | 66 | TTC | -1 | ||
| tRNAPhe | 7 096-7 163 | N | 68 | GAA | -3 | ||
| nad5 | 7 162-8 878 | N | 1 717 | ATT | TAA | -2 | |
| tRNAHis | 8 881-8 944 | N | 64 | GTG | 2 | ||
| nad4 | 8 943-10 280 | N | 1 338 | ATG | TA(A) | -2 | |
| nad4l | 10 273-10 606 | N | 334 | TTG | TAA | -8 | |
| tRNAThr | 10 566-10 631 | J | 66 | TGT | -41 | ||
| tRNAPro | 10 631-10 696 | N | 66 | TGG | -1 | ||
| nad6 | 10 697-11 201 | J | 505 | ATC | TAA | 0 | |
| cytb | 11 200-12 343 | J | 1 144 | ATG | TAG | -2 | |
| tRNASer(UCN) | 12 341-12 406 | J | 66 | TGA | -3 | ||
| nad1 | 12 422-13 370 | N | 949 | ATA | TAG | 15 | |
| tRNALeu(CUN) | 13 374-13 440 | N | 67 | TAG | 3 | ||
| 16S rRNA | 13 404-14 753 | N | 1 350 | -37 | |||
| tRNAVal | 14 731-14 801 | N | 71 | TAC | -23 | ||
| 12S rRNA | 14 801-15 599 | N | 800 | -1 | |||
| Control Region | 15 600-16 673 | J | 1 074 |
| 基因Gene | A/% | T/% | G/% | C/% | (A+T)/% | (G+C)/% | AT-skew | GC-skew |
|---|---|---|---|---|---|---|---|---|
| 全基因组 Whole genome | 38.97 | 36.89 | 9.21 | 14.93 | 75.87 | 24.13 | 0.027 | -0.237 |
| 13PCGs | 32.16 | 42.71 | 12.25 | 12.88 | 74.87 | 25.13 | -0.141 | -0.025 |
| 22tRNAs | 38.79 | 37.75 | 13.46 | 10.01 | 76.54 | 23.46 | 0.014 | 0.147 |
| 2rRNAs | 36.41 | 39.39 | 16.76 | 7.45 | 75.79 | 24.21 | -0.039 | 0.385 |
| Control region | 41.99 | 39.01 | 4.19 | 14.80 | 81.01 | 18.99 | 0.037 | -0.559 |
| 16S rRNA | 36.84 | 40.85 | 15.64 | 6.67 | 77.69 | 22.31 | 0.052 | 0.402 |
| 12S rRNA | 35.67 | 36.92 | 18.65 | 8.76 | 72.59 | 27.41 | -0.017 | 0.361 |
| nad2 | 35.52 | 41.27 | 7.04 | 16.17 | 76.79 | 23.21 | -0.075 | -0.393 |
| cox1 | 31.26 | 37.35 | 14.24 | 17.15 | 68.61 | 31.39 | -0.089 | -0.093 |
| cox2 | 34.32 | 38.14 | 11.72 | 15.82 | 72.46 | 27.54 | -0.053 | -0.149 |
| atp8 | 39.10 | 37.82 | 8.33 | 14.74 | 76.92 | 23.08 | 0.017 | -0.278 |
| atp6 | 31.99 | 42.26 | 11.16 | 14.58 | 74.26 | 25.74 | -0.138 | -0.133 |
| cox3 | 31.22 | 40.23 | 13.07 | 15.48 | 71.45 | 28.55 | -0.127 | -0.084 |
| nad3 | 32.20 | 44.07 | 9.89 | 13.84 | 76.27 | 23.73 | -0.158 | -0.166 |
| nad5 | 31.59 | 45.98 | 13.29 | 9.15 | 77.56 | 22.44 | -0.186 | 0.184 |
| nad4 | 30.96 | 46.67 | 14.06 | 8.30 | 77.64 | 22.36 | -0.202 | 0.258 |
| nad4l | 30.33 | 50.75 | 13.21 | 5.71 | 81.08 | 18.92 | -0.252 | 0.396 |
| nad6 | 37.50 | 43.45 | 6.55 | 12.50 | 80.95 | 19.05 | -0.074 | -0.312 |
| cytb | 32.46 | 39.63 | 11.90 | 16.01 | 72.09 | 27.91 | -0.099 | -0.147 |
| nad1 | 28.38 | 48.00 | 15.30 | 8.33 | 76.37 | 23.63 | -0.257 | 0.295 |
表2 铜绿丽金龟线粒体基因组碱基组成
Table 2 Base composition in the mitochondrial genome of A. corpulenta
| 基因Gene | A/% | T/% | G/% | C/% | (A+T)/% | (G+C)/% | AT-skew | GC-skew |
|---|---|---|---|---|---|---|---|---|
| 全基因组 Whole genome | 38.97 | 36.89 | 9.21 | 14.93 | 75.87 | 24.13 | 0.027 | -0.237 |
| 13PCGs | 32.16 | 42.71 | 12.25 | 12.88 | 74.87 | 25.13 | -0.141 | -0.025 |
| 22tRNAs | 38.79 | 37.75 | 13.46 | 10.01 | 76.54 | 23.46 | 0.014 | 0.147 |
| 2rRNAs | 36.41 | 39.39 | 16.76 | 7.45 | 75.79 | 24.21 | -0.039 | 0.385 |
| Control region | 41.99 | 39.01 | 4.19 | 14.80 | 81.01 | 18.99 | 0.037 | -0.559 |
| 16S rRNA | 36.84 | 40.85 | 15.64 | 6.67 | 77.69 | 22.31 | 0.052 | 0.402 |
| 12S rRNA | 35.67 | 36.92 | 18.65 | 8.76 | 72.59 | 27.41 | -0.017 | 0.361 |
| nad2 | 35.52 | 41.27 | 7.04 | 16.17 | 76.79 | 23.21 | -0.075 | -0.393 |
| cox1 | 31.26 | 37.35 | 14.24 | 17.15 | 68.61 | 31.39 | -0.089 | -0.093 |
| cox2 | 34.32 | 38.14 | 11.72 | 15.82 | 72.46 | 27.54 | -0.053 | -0.149 |
| atp8 | 39.10 | 37.82 | 8.33 | 14.74 | 76.92 | 23.08 | 0.017 | -0.278 |
| atp6 | 31.99 | 42.26 | 11.16 | 14.58 | 74.26 | 25.74 | -0.138 | -0.133 |
| cox3 | 31.22 | 40.23 | 13.07 | 15.48 | 71.45 | 28.55 | -0.127 | -0.084 |
| nad3 | 32.20 | 44.07 | 9.89 | 13.84 | 76.27 | 23.73 | -0.158 | -0.166 |
| nad5 | 31.59 | 45.98 | 13.29 | 9.15 | 77.56 | 22.44 | -0.186 | 0.184 |
| nad4 | 30.96 | 46.67 | 14.06 | 8.30 | 77.64 | 22.36 | -0.202 | 0.258 |
| nad4l | 30.33 | 50.75 | 13.21 | 5.71 | 81.08 | 18.92 | -0.252 | 0.396 |
| nad6 | 37.50 | 43.45 | 6.55 | 12.50 | 80.95 | 19.05 | -0.074 | -0.312 |
| cytb | 32.46 | 39.63 | 11.90 | 16.01 | 72.09 | 27.91 | -0.099 | -0.147 |
| nad1 | 28.38 | 48.00 | 15.30 | 8.33 | 76.37 | 23.63 | -0.257 | 0.295 |
| 氨基酸Amino acid | 密码子Codon | 使用次数Usage frequency | RSCU | 氨基酸Amino acid | 密码子Codon | 使用次数Usage frequency | RSCU | |
|---|---|---|---|---|---|---|---|---|
| Phe(F) | UUU | 322 | 1.62 | Pro(P) | CCU | 39 | 1.73 | |
| UUC | 75 | 0.38 | CCC | 27 | 1.20 | |||
| Leu(L) | UUA | 258 | 2.92 | CCA | 22 | 0.98 | ||
| UUG | 53 | 0.60 | CCG | 2 | 0.09 | |||
| CUU | 108 | 1.22 | Thr(T) | ACU | 74 | 1.72 | ||
| CUC | 20 | 0.23 | ACC | 21 | 0.49 | |||
| CUA | 72 | 0.82 | ACA | 67 | 1.56 | |||
| CUG | 19 | 0.22 | ACG | 10 | 0.23 | |||
| Ile(I) | AUU | 287 | 1.69 | Ala(A) | GCU | 52 | 2.29 | |
| AUC | 52 | 0.31 | GCC | 12 | 0.53 | |||
| Met(M) | AUA | 164 | 1.55 | GCA | 25 | 1.10 | ||
| AUG | 47 | 0.45 | GCG | 2 | 0.09 | |||
| Val(V) | GUU | 72 | 1.81 | Tyr(Y) | UAU | 212 | 1.51 | |
| GUC | 13 | 0.33 | UAC | 68 | 0.49 | |||
| GUA | 56 | 1.41 | His(H) | CAU | 69 | 1.73 | ||
| GUG | 18 | 0.45 | CAC | 11 | 0.28 | |||
| Ser(S) | UCU | 69 | 1.71 | Gln(Q) | CAA | 56 | 1.58 | |
| UCC | 25 | 0.62 | CAG | 15 | 0.42 | |||
| UCA | 56 | 1.39 | Asn(N) | AAU | 147 | 1.47 | ||
| UCG | 9 | 0.22 | AAC | 53 | 0.53 | |||
| AGU | 50 | 1.24 | Lys(K) | AAA | 84 | 1.47 | ||
| AGC | 21 | 0.52 | AAG | 30 | 0.53 | |||
| AGA | 58 | 1.44 | Asp(D) | GAU | 53 | 1.58 | ||
| AGG | 34 | 0.84 | GAC | 14 | 0.42 | |||
| Glu(E) | GAA | 57 | 1.50 | Cys(C) | UGU | 34 | 1.31 | |
| GAG | 19 | 0.50 | UGC | 18 | 0.69 | |||
| Trp(W) | UGA | 75 | 1.53 | Gly(G) | GGU | 29 | 0.91 | |
| UGG | 23 | 0.47 | GGC | 9 | 0.28 | |||
| Arg(R) | CGU | 10 | 1.25 | GGA | 70 | 2.19 | ||
| CGC | 1 | 0.13 | GGG | 20 | 0.63 | |||
| CGA | 19 | 2.38 | * | UAA* | 152 | 1.33 | ||
| CGG | 2 | 0.25 | UAG* | 76 | 0.67 |
表3 铜绿丽金龟线粒体基因组相对同义密码子使用频率(RSCU)
Table 3 Relative synonymous codon usage(RSCU)in the mitochondrial genome of A. corpulenta
| 氨基酸Amino acid | 密码子Codon | 使用次数Usage frequency | RSCU | 氨基酸Amino acid | 密码子Codon | 使用次数Usage frequency | RSCU | |
|---|---|---|---|---|---|---|---|---|
| Phe(F) | UUU | 322 | 1.62 | Pro(P) | CCU | 39 | 1.73 | |
| UUC | 75 | 0.38 | CCC | 27 | 1.20 | |||
| Leu(L) | UUA | 258 | 2.92 | CCA | 22 | 0.98 | ||
| UUG | 53 | 0.60 | CCG | 2 | 0.09 | |||
| CUU | 108 | 1.22 | Thr(T) | ACU | 74 | 1.72 | ||
| CUC | 20 | 0.23 | ACC | 21 | 0.49 | |||
| CUA | 72 | 0.82 | ACA | 67 | 1.56 | |||
| CUG | 19 | 0.22 | ACG | 10 | 0.23 | |||
| Ile(I) | AUU | 287 | 1.69 | Ala(A) | GCU | 52 | 2.29 | |
| AUC | 52 | 0.31 | GCC | 12 | 0.53 | |||
| Met(M) | AUA | 164 | 1.55 | GCA | 25 | 1.10 | ||
| AUG | 47 | 0.45 | GCG | 2 | 0.09 | |||
| Val(V) | GUU | 72 | 1.81 | Tyr(Y) | UAU | 212 | 1.51 | |
| GUC | 13 | 0.33 | UAC | 68 | 0.49 | |||
| GUA | 56 | 1.41 | His(H) | CAU | 69 | 1.73 | ||
| GUG | 18 | 0.45 | CAC | 11 | 0.28 | |||
| Ser(S) | UCU | 69 | 1.71 | Gln(Q) | CAA | 56 | 1.58 | |
| UCC | 25 | 0.62 | CAG | 15 | 0.42 | |||
| UCA | 56 | 1.39 | Asn(N) | AAU | 147 | 1.47 | ||
| UCG | 9 | 0.22 | AAC | 53 | 0.53 | |||
| AGU | 50 | 1.24 | Lys(K) | AAA | 84 | 1.47 | ||
| AGC | 21 | 0.52 | AAG | 30 | 0.53 | |||
| AGA | 58 | 1.44 | Asp(D) | GAU | 53 | 1.58 | ||
| AGG | 34 | 0.84 | GAC | 14 | 0.42 | |||
| Glu(E) | GAA | 57 | 1.50 | Cys(C) | UGU | 34 | 1.31 | |
| GAG | 19 | 0.50 | UGC | 18 | 0.69 | |||
| Trp(W) | UGA | 75 | 1.53 | Gly(G) | GGU | 29 | 0.91 | |
| UGG | 23 | 0.47 | GGC | 9 | 0.28 | |||
| Arg(R) | CGU | 10 | 1.25 | GGA | 70 | 2.19 | ||
| CGC | 1 | 0.13 | GGG | 20 | 0.63 | |||
| CGA | 19 | 2.38 | * | UAA* | 152 | 1.33 | ||
| CGG | 2 | 0.25 | UAG* | 76 | 0.67 |

图3 基于线粒体蛋白质编码基因核苷酸序列构建的铜绿丽金龟与其他金龟科昆虫的系统发育树(最大似然法和贝叶斯法) 进化枝上方为置信值,下方为遗传距离;斜杠前后分别代表最大似然法和贝叶斯法计算的结果
Fig. 3 Phylogenetic tree of A. corpulenta and other insects species of Scarabaeidae based on the mitochondrial protein-coding gene sequences(maximum likelihood and Bayesian) The values on top and bottom of the branches refer to the confidence values and genetic distance, respectively. The values before and after the slash refer to the results of the maximum likelihood estimation and Bayesian algorithm, respectively
| P. brevitarsis | Adoretus sp. | A. corpulenta | C. jansoni | E. chinensis | H. oblita | H. parallela | Leucocelis sp. | M. splendens | O. rhinoceros | O. opicum | P. laticollis | P. japonica | P. mutans | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Protaetia brevitarsis | ||||||||||||||
| Adoretus sp. | 0.2334 | |||||||||||||
| Anomala corpulenta | 0.2371 | 0.2099 | ||||||||||||
| Cheirotonus jansoni | 0.3535 | 0.3507 | 0.3494 | |||||||||||
| Eophileurus chinensis | 0.3131 | 0.2969 | 0.3025 | 0.3696 | ||||||||||
| Holotrichia oblita | 0.2868 | 0.2723 | 0.2785 | 0.3598 | 0.3447 | |||||||||
| Holotrichia parallela | 0.2570 | 0.2515 | 0.2570 | 0.3486 | 0.3300 | 0.2964 | ||||||||
| Leucocelis sp. | 0.1669 | 0.2319 | 0.2314 | 0.3403 | 0.3051 | 0.2899 | 0.2614 | |||||||
| Mimela splendens | 0.2380 | 0.2090 | 0.0079 | 0.3502 | 0.3022 | 0.2795 | 0.2557 | 0.2307 | ||||||
| Oryctes rhinoceros | 0.3025 | 0.2726 | 0.2842 | 0.3559 | 0.2574 | 0.3219 | 0.3051 | 0.2927 | 0.2839 | |||||
| Osmoderma opicum | 0.2056 | 0.2803 | 0.2758 | 0.3344 | 0.3460 | 0.3193 | 0.2929 | 0.2386 | 0.2762 | 0.3362 | ||||
| Polyphylla laticollis | 0.3173 | 0.3015 | 0.3172 | 0.3798 | 0.3607 | 0.3543 | 0.2968 | 0.3233 | 0.3169 | 0.3513 | 0.3266 | |||
| Popillia japonica | 0.2522 | 0.2218 | 0.1774 | 0.3615 | 0.3100 | 0.2948 | 0.2720 | 0.2485 | 0.1779 | 0.2919 | 0.2870 | 0.3122 | ||
| Popillia mutans | 0.2472 | 0.2190 | 0.1727 | 0.3562 | 0.3132 | 0.2901 | 0.2651 | 0.2477 | 0.1735 | 0.2875 | 0.2902 | 0.3159 | 0.1578 | |
| Rhopaea magnicornis | 0.2553 | 0.2382 | 0.2445 | 0.3542 | 0.3219 | 0.2937 | 0.2362 | 0.2521 | 0.2461 | 0.3011 | 0.2563 | 0.2897 | 0.2588 | 0.2521 |
表4 铜绿丽金龟与其他植食性金龟科昆虫的线粒体蛋白质编码基因基于Kimura-2-Parameter参数的遗传距离
Table 4 Pairwise genetic distances of mitochondrial protein-coding gene sequences between A. corpulenta and other phytophagous species of Scarabaeidae based on Kimura-2-Parameters
| P. brevitarsis | Adoretus sp. | A. corpulenta | C. jansoni | E. chinensis | H. oblita | H. parallela | Leucocelis sp. | M. splendens | O. rhinoceros | O. opicum | P. laticollis | P. japonica | P. mutans | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Protaetia brevitarsis | ||||||||||||||
| Adoretus sp. | 0.2334 | |||||||||||||
| Anomala corpulenta | 0.2371 | 0.2099 | ||||||||||||
| Cheirotonus jansoni | 0.3535 | 0.3507 | 0.3494 | |||||||||||
| Eophileurus chinensis | 0.3131 | 0.2969 | 0.3025 | 0.3696 | ||||||||||
| Holotrichia oblita | 0.2868 | 0.2723 | 0.2785 | 0.3598 | 0.3447 | |||||||||
| Holotrichia parallela | 0.2570 | 0.2515 | 0.2570 | 0.3486 | 0.3300 | 0.2964 | ||||||||
| Leucocelis sp. | 0.1669 | 0.2319 | 0.2314 | 0.3403 | 0.3051 | 0.2899 | 0.2614 | |||||||
| Mimela splendens | 0.2380 | 0.2090 | 0.0079 | 0.3502 | 0.3022 | 0.2795 | 0.2557 | 0.2307 | ||||||
| Oryctes rhinoceros | 0.3025 | 0.2726 | 0.2842 | 0.3559 | 0.2574 | 0.3219 | 0.3051 | 0.2927 | 0.2839 | |||||
| Osmoderma opicum | 0.2056 | 0.2803 | 0.2758 | 0.3344 | 0.3460 | 0.3193 | 0.2929 | 0.2386 | 0.2762 | 0.3362 | ||||
| Polyphylla laticollis | 0.3173 | 0.3015 | 0.3172 | 0.3798 | 0.3607 | 0.3543 | 0.2968 | 0.3233 | 0.3169 | 0.3513 | 0.3266 | |||
| Popillia japonica | 0.2522 | 0.2218 | 0.1774 | 0.3615 | 0.3100 | 0.2948 | 0.2720 | 0.2485 | 0.1779 | 0.2919 | 0.2870 | 0.3122 | ||
| Popillia mutans | 0.2472 | 0.2190 | 0.1727 | 0.3562 | 0.3132 | 0.2901 | 0.2651 | 0.2477 | 0.1735 | 0.2875 | 0.2902 | 0.3159 | 0.1578 | |
| Rhopaea magnicornis | 0.2553 | 0.2382 | 0.2445 | 0.3542 | 0.3219 | 0.2937 | 0.2362 | 0.2521 | 0.2461 | 0.3011 | 0.2563 | 0.2897 | 0.2588 | 0.2521 |
| [1] | 鞠倩, 郭晓强, 李晓, 等. 铜绿丽金龟对寄主植物挥发物的触角电生理及行为反应[J]. 植物保护学报, 2016, 43(2): 281-287. |
| Ju Q, Guo XQ, Li X, et al. EAG and behavioral responses of copper-green chafer Anomala corpulenta Motschulsky(Coleoptera: Rutelidae)[J]. J Plant Prot, 2016, 43(2): 281-287. | |
| [2] | Li X, Ju Q, Jie WC, et al. Chemosensory gene families in adult antennae of Anomala corpulenta Motschulsky(Coleoptera: Scarabaeidae: Rutelinae)[J]. PLoS One, 2015, 10(4): e0121504. |
| [3] |
Bouchard P, Bousquet Y, Davies AE, et al. Family-group names in Coleoptera(Insecta)[J]. ZooKeys, 2011(88): 1-972.
doi: 10.3897/zookeys.88.807 pmid: 21594053 |
| [4] | 路园园, 丁强, 杨星科, 等. 中国丽金龟亚科昆虫分类学研究进展[J]. 环境昆虫学报, 2022. http://kns.cnki.net/kcms/detail/44.1640.Q.20220509.1919.002.html. |
| Lu YY, Ding Q, Yang XK, et al. Advances in taxonomy of Chinese Rutelinae[J]. J Environ Entomol, 2022. http://kns.cnki.net/kcms/detail/44.1640.Q.20220509.1919.002.html. | |
| [5] |
Cameron SL. Insect mitochondrial genomics: implications for evolution and phylogeny[J]. Annu Rev Entomol, 2014, 59: 95-117.
doi: 10.1146/annurev-ento-011613-162007 pmid: 24160435 |
| [6] |
Wei SJ, Shi M, Chen XX, et al. New views on strand asymmetry in insect mitochondrial genomes[J]. PLoS One, 2010, 5(9): e12708.
doi: 10.1371/journal.pone.0012708 URL |
| [7] | 方晨晨, 郭晓华, 刘广纯, 等. 基因序列在金龟总科(Scarabaeoidea)分子系统学研究中的应用[J]. 应用昆虫学报, 2012, 49(4): 1048-1055. |
| Fang CC, Guo XH, Liu GC, et al. Application of gene sequences to the molecular systematics of the Scarabaeoidea[J]. Chin J Appl Entomol, 2012, 49(4): 1048-1055. | |
| [8] | 申欣, 李晓, 徐启华. 日本鼓虾与鲜明鼓虾线粒体基因组全序列的分析比较[J]. 海洋学报, 2012, 34(5): 147-153. |
|
Shen X, Li X, Xu QH. Comparison and analysis of Alpheus japonicus and A. distinguendus complete mitochondrial genome sequences[J]. Acta Oceanol Sin, 2012, 34(5): 147-153.
doi: 10.1007/s13131-015-0766-9 URL |
|
| [9] |
薛清, 杜虹锐, 薛会英, 等. 苜蓿滑刃线虫线粒体基因组及其系统发育研究[J]. 生物技术通报, 2021, 37(7): 98-106.
doi: 10.13560/j.cnki.biotech.bull.1985.2021-0628 |
| Xue Q, Du HR, Xue HY, et al. Mitochondrial genome and phylogeny of Aphelenchoides medicagus[J]. Biotechnol Bull, 2021, 37(7): 98-106. | |
| [10] |
Ye F, Samuels DC, Clark T, et al. High-throughput sequencing in mitochondrial DNA research[J]. Mitochondrion, 2014, 17: 157-163.
doi: 10.1016/j.mito.2014.05.004 pmid: 24859348 |
| [11] |
Yang WZ, Zhang Y, Feng SQ, et al. The first complete mitochondrial genome of the Japanese beetle Popillia japonica(Coleoptera: Scarabaeidae)and its phylogenetic implications for the superfamily Scarabaeoidea[J]. Int J Biol Macromol, 2018, 118(Pt B): 1406-1413.
doi: 10.1016/j.ijbiomac.2018.06.131 URL |
| [12] | Song N, Zhang H. The mitochondrial genomes of phytophagous scarab beetles and systematic implications[J]. J Insect Sci, 2018, 18(6): 11. |
| [13] | 万书波. 中国花生栽培学[M]. 上海: 上海科学技术出版社, 2003. |
| Wan SB. Peanut cultivation in China[M]. Shanghai: Shanghai Scientific & Technical Publishers, 2003. | |
| [14] | 张鸿飞. 蓖麻对华北地区关键金龟甲种类趋性、交配和解毒代谢的影响[D]. 郑州: 河南农业大学, 2018. |
| Zhang HF. Effects of Castor bean on taxis, mating, and detoxification of key scarab species in North China[D]. Zhengzhou: Henan Agricultural University, 2018. | |
| [15] |
Patel RK, Jain M. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data[J]. PLoS One, 2012, 7(2): e30619.
doi: 10.1371/journal.pone.0030619 URL |
| [16] |
Bankevich A, Nurk S, Antipov D, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing[J]. J Comput Biol, 2012, 19(5): 455-477.
doi: 10.1089/cmb.2012.0021 pmid: 22506599 |
| [17] |
Boetzer M, Henkel CV, Jansen HJ, et al. Scaffolding pre-assembled contigs using SSPACE[J]. Bioinformatics, 2011, 27(4): 578-579.
doi: 10.1093/bioinformatics/btq683 pmid: 21149342 |
| [18] |
Bernt M, Donath A, Jühling F, et al. MITOS: improved de novo metazoan mitochondrial genome annotation[J]. Mol Phylogenet Evol, 2013, 69(2): 313-319.
doi: 10.1016/j.ympev.2012.08.023 URL |
| [19] |
Lowe TM, Chan PP. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes[J]. Nucleic Acids Res, 2016, 44(W1): W54-W57.
doi: 10.1093/nar/gkw413 URL |
| [20] |
Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11[J]. Mol Biol Evol, 2021, 38(7): 3022-3027.
doi: 10.1093/molbev/msab120 pmid: 33892491 |
| [21] |
Larkin MA, Blackshields G, Brown NP, et al. Clustal W and Clustal X version 2.0[J]. Bioinformatics, 2007, 23(21): 2947-2948.
doi: 10.1093/bioinformatics/btm404 pmid: 17846036 |
| [22] |
Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis[J]. Mol Biol Evol, 2000, 17(4): 540-552.
doi: 10.1093/oxfordjournals.molbev.a026334 pmid: 10742046 |
| [23] |
Vaidya G, Lohman DJ, Meier R. SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and Codon information[J]. Cladistics, 2011, 27(2): 171-180.
doi: 10.1111/j.1096-0031.2010.00329.x URL |
| [24] |
Lefort V, Longueville JE, Gascuel O. SMS: smart model selection in PhyML[J]. Mol Biol Evol, 2017, 34(9): 2422-2424.
doi: 10.1093/molbev/msx149 pmid: 28472384 |
| [25] |
Kalyaanamoorthy S, Minh BQ, Wong TKF, et al. ModelFinder: fast model selection for accurate phylogenetic estimates[J]. Nat Methods, 2017, 14(6): 587-589.
doi: 10.1038/nmeth.4285 pmid: 28481363 |
| [26] |
Guindon S, Dufayard JF, Lefort V, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0[J]. Syst Biol, 2010, 59(3): 307-321.
doi: 10.1093/sysbio/syq010 pmid: 20525638 |
| [27] |
Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models[J]. Bioinformatics, 2003, 19(12): 1572-1574.
doi: 10.1093/bioinformatics/btg180 pmid: 12912839 |
| [28] |
Ojala D, Montoya J, Attardi G. tRNA punctuation model of RNA processing in human mitochondria[J]. Nature, 1981, 290(5806): 470-474.
doi: 10.1038/290470a0 URL |
| [29] | Liao F, Wang L, Wu S, et al. The complete mitochondrial genome of the fall webworm, Hyphantria cunea(Lepidoptera: Arctiidae)[J]. Int J Biol Sci, 2010, 6(2): 172-186. |
| [30] |
Wolstenholme DR. Animal mitochondrial DNA: structure and evolution[J]. Int Rev Cytol, 1992, 141: 173-216.
pmid: 1452431 |
| [31] |
Clary DO, Wolstenholme DR. The mitochondrial DNA molecular of Drosophila yakuba: nucleotide sequence, gene organization, and genetic code[J]. J Mol Evol, 1985, 22(3): 252-271.
pmid: 3001325 |
| [32] |
Bohacz C, James GH, Ahrens D. Comparative morphology of antennal surface structures in pleurostict scarab beetles(Coleoptera)[J]. Zoomorphology, 2020, 139(3): 327-346.
doi: 10.1007/s00435-020-00495-0 URL |
| [33] |
Ayivi SPG, Tong Y, Storey KB, et al. The mitochondrial genomes of 18 new pleurosticti(Coleoptera: Scarabaeidae)exhibit a novel trnQ-NCR-trnI-trnM gene rearrangement and clarify phylogenetic relationships of subfamilies within Scarabaeidae[J]. Insects, 2021, 12(11): 1025.
doi: 10.3390/insects12111025 URL |
| [34] |
Ahrens D. The phylogeny of Sericini and their position within the Scarabaeidae based on morphological characters(Coleoptera: Scarabaeidae)[J]. Syst Entomol, 2006, 31(1): 113-144.
doi: 10.1111/j.1365-3113.2005.00307.x URL |
| [35] |
Coca-Abia MM. Phylogenetic relationships of the subfamily melolonthinae(Coleoptera, Scarabaeidae)[J]. Insect Syst Evol, 2007, 38(4): 447-472.
doi: 10.1163/187631207794760921 URL |
| [1] | 尹明华, 余锾媛, 肖心怡, 王玉婷. 江西铅山红芽芋叶绿体基因组特征及系统发育分析[J]. 生物技术通报, 2023, 39(6): 233-247. |
| [2] | 刘雄伟, 刘畅, 曾宪法, 杨小英, 俸婷婷, 赵杰宏, 周英. 朱砂根叶绿体全基因组解析及系统发育分析[J]. 生物技术通报, 2023, 39(1): 232-242. |
| [3] | 刘警鞠, 张雨森, 陈娟, 孙炳达, 赵国柱. 曲霉属的现代分类命名研究进展[J]. 生物技术通报, 2022, 38(7): 109-118. |
| [4] | 钱方, 高作敏, 胡利娟, 王洪程. 海甘蓝(Crambe abyssinica)叶绿体基因组特征及其系统发育研究[J]. 生物技术通报, 2022, 38(6): 174-186. |
| [5] | 程赛赛, 龚鑫, 薛文凤, 万兵兵, 刘满强, 胡锋. 生态进化视角的植物-微生物组互作研究进展[J]. 生物技术通报, 2020, 36(9): 3-13. |
| [6] | 李裕华, 任永康, 赵兴华, 刘江, 韩斌, 王长彪, 唐朝晖. 禾本科主要农作物叶绿体基因组研究进展[J]. 生物技术通报, 2020, 36(11): 112-121. |
| [7] | 刘凌燕, 陈志宇, 曾还雄, 林培彬, 金小宝. 美洲大蠊肠道内生微杆菌的分离鉴定及其抑菌活性研究[J]. 生物技术通报, 2018, 34(6): 172-177. |
| [8] | 张广志, 王加宁, 吴晓青, 周方园, 张新建, 赵晓燕, 谢雪迎, 周红姿. 设施番茄根围土样中木霉菌多样性及功能活性分析[J]. 生物技术通报, 2018, 34(4): 179-185. |
| [9] | 胡文哲, 谭泽文, 王勇, 徐羡微, 谭志远. 藤县药用野生稻内生固氮菌分离鉴定及系统发育分析[J]. 生物技术通报, 2016, 32(6): 111-119. |
| [10] | 李丽卡, 李象钦, 谢国文, 李海生, 郑毅胜, 藤婕华, 高锦伟. 基于cpDNA片段探讨中-日间断分布双花木属植物的系统发育学[J]. 生物技术通报, 2016, 32(1): 80-87. |
| [11] | 刘国红, 刘波, 林营志, 唐建阳. 基于脂肪酸生物标记与16S rRNA的芽胞杆菌系统发育分析比较[J]. 生物技术通报, 2015, 31(3): 146-153. |
| [12] | 焦林培金,黄运红,李素珍,龙中儿. 一株拮抗链霉菌的鉴定及其多相分类特征[J]. 生物技术通报, 2015, 31(1): 138-143. |
| [13] | 陈秀玲, 李景富, 张丽莉, 张俊峰, 王傲雪. 一株淡色生赤壳菌的生防作用分析及系统发育树构建[J]. 生物技术通报, 2014, 0(5): 184-189. |
| [14] | 丁玫, 陈祥, 许厚强, 吴通贵, 冯文武, 柏琳. 贵州三个地方猪种的系统发育分析[J]. 生物技术通报, 2014, 0(4): 96-101. |
| [15] | 王菲, 刘抗, 范家同, 成守亮, 王子元. 一株耐盐弧菌Vibrio sp.K1-L的分离与鉴定[J]. 生物技术通报, 2014, 0(4): 127-131. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||