








生物技术通报 ›› 2022, Vol. 38 ›› Issue (6): 174-186.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1078
收稿日期:2021-08-21
出版日期:2022-06-26
发布日期:2022-07-11
作者简介:钱方,男,硕士研究生,研究方向:油菜远缘杂交及种质资源创新;E-mail: 基金资助:
QIAN Fang( ), GAO Zuo-min, HU Li-juan, WANG Hong-cheng(
), GAO Zuo-min, HU Li-juan, WANG Hong-cheng( )
)
Received:2021-08-21
Published:2022-06-26
Online:2022-07-11
摘要:
海甘蓝(Crambe abyssinica)为十字花科(Brassicaceae)海甘蓝属(Crambe)植物,其种子富含芥酸,被视为一种重要的可再生工业油用资源。本研究基于Illumina HiSeq 测序数据从头组装了(de novo)海甘蓝的叶绿体(chloroplast,cp)基因组,并分析其在十字花科植物中的系统发育位置。结果表明,海甘蓝叶绿体基因组大小为153 754 bp,呈典型的四分体结构;其包含111个基因,包括78个编码基因、29个tRNA基因以及4个rRNA基因。通过密码子偏好性分析,海甘蓝叶绿体基因组中末端为A/U的密码子使用存在偏倚。SSR及长重复片段分析检测到41个重复序列和59个SSR位点。与近缘种比较,发现其叶绿体基因组序列高度保守,尤其蛋白质编码序列相似度极高。同义与非同义核苷酸替代率分析表明,除ndhB和ycf2基因外,其余编码基因普遍存在纯化选择压力。此外,系统发育分析发现海甘蓝与芸薹属重要的经济作物亲缘关系密切,与白芥属植物形成姊妹类群。这为海甘蓝与芸薹属植物的种间杂交提供了遗传支撑,同时也为阐明海甘蓝的进化史提供了重要的分子信息。
钱方, 高作敏, 胡利娟, 王洪程. 海甘蓝(Crambe abyssinica)叶绿体基因组特征及其系统发育研究[J]. 生物技术通报, 2022, 38(6): 174-186.
QIAN Fang, GAO Zuo-min, HU Li-juan, WANG Hong-cheng. Characteristics of Crambe abyssinica Chloroplast Genome and Its Phylogenetic Relationship in Brassicaceae[J]. Biotechnology Bulletin, 2022, 38(6): 174-186.
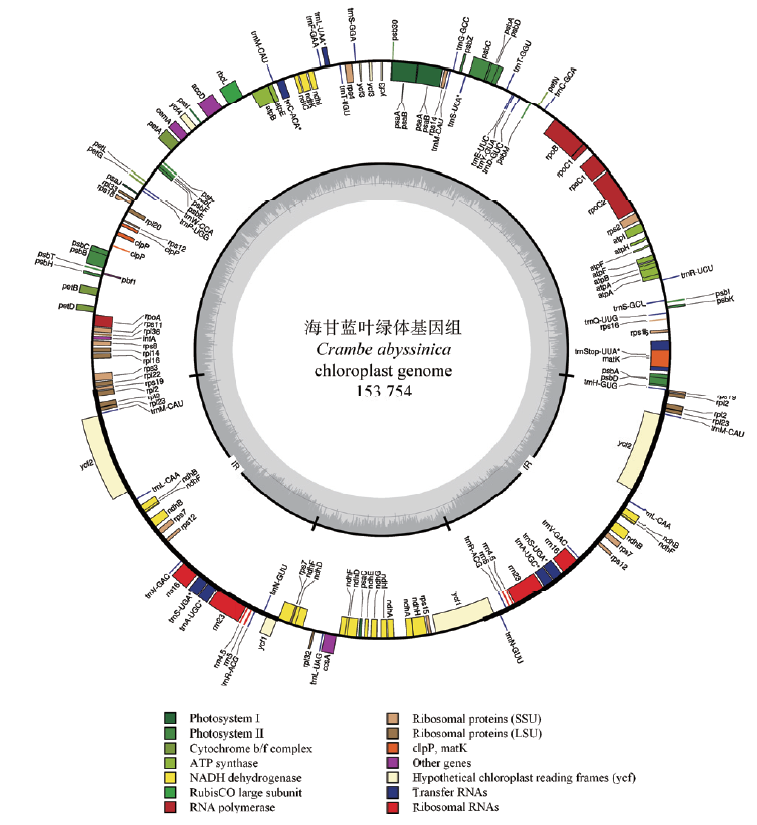
图1 海甘蓝叶绿体全基因组图谱 圆圈外面的基因是顺时针转录,而圆圈里面的基因是逆时针转录。不同功能组的基因以不同的颜色显示。内圈深灰色对应GC含量,浅灰色对应AT含量
Fig.1 Circle gene map of the C. abyssinica cp genome Genes drawn outside and inside of the circle are transcribed clockwise and counterclockwise,respectively. Genes belonging to different functional groups are color coded. The dark gray in the inner circle corresponds to GC content,and light grey to AT content
| 类别 Category | 条目 Item | 说明 Description |
|---|---|---|
| 叶绿体基因组结构 | 大单拷贝区/bp | 83 622 |
| Structure of cp genome | 反向重复区/bp | 26 176 |
| 小单拷贝区/bp | 17 780 | |
| 基因组大小/bp | 153 754 | |
| 基因组成 | 基因总数 | 111 |
| Gene composition | 编码基因 | 78 |
| tRNAs | 29 | |
| rRNAs | 4 | |
| 双拷贝基因 | 19 | |
| 大单拷贝区基因数 | 81 | |
| 反向重复区基因数 | 18 | |
| 小单拷贝区基因数 | 13 | |
| GC含量 | 大单拷贝区GC含量/% | 34.14 |
| GC content | 反向重复区GC含量/% | 42.36 |
| 小单拷贝区GC含量/% | 29.29 | |
| 平均GC含量/% | 36.37 | |
| 单碱基含量 | A/% | 31.35 |
| Single nucleotide content | T/% | 32.28 |
| C/% | 18.51 | |
| G/% | 17.86 |
表1 海甘蓝叶绿体基因组的详细特征
Table 1 Detailed characteristics of the chloroplast genome of C. abyssinica
| 类别 Category | 条目 Item | 说明 Description |
|---|---|---|
| 叶绿体基因组结构 | 大单拷贝区/bp | 83 622 |
| Structure of cp genome | 反向重复区/bp | 26 176 |
| 小单拷贝区/bp | 17 780 | |
| 基因组大小/bp | 153 754 | |
| 基因组成 | 基因总数 | 111 |
| Gene composition | 编码基因 | 78 |
| tRNAs | 29 | |
| rRNAs | 4 | |
| 双拷贝基因 | 19 | |
| 大单拷贝区基因数 | 81 | |
| 反向重复区基因数 | 18 | |
| 小单拷贝区基因数 | 13 | |
| GC含量 | 大单拷贝区GC含量/% | 34.14 |
| GC content | 反向重复区GC含量/% | 42.36 |
| 小单拷贝区GC含量/% | 29.29 | |
| 平均GC含量/% | 36.37 | |
| 单碱基含量 | A/% | 31.35 |
| Single nucleotide content | T/% | 32.28 |
| C/% | 18.51 | |
| G/% | 17.86 |
| 基因功能分类 Category for genes | 基因功能 Groups of genes | 基因名 Names of genes |
|---|---|---|
| 参与光合作用的基因 Genes involving in photosynthesis | 光合系统亚基 | ndhI,ndhJ,ndhK,psaA,psaB,psaC,psaI,psaJ,psbA,psbB,psbC,psbD,psbE,psbF,psbH,psbI,psbJ,psbK,psbL,psbM,psbN,psbT,psbZ |
| 细胞色素复合体亚基 | petA,petBb,petDb,petG,petL,petN | |
| 二磷酸核酮糖羧化酶大亚基 | rbcL | |
| ATP合成酶亚基 | atpA,atpB,atpE,atpFb,atpH,atpI | |
| NADH脱氢酶亚基 | ndhAb,ndhBa,b,ndhC,ndhD,ndhE,ndhF,ndhG,ndhH | |
| 自身复制相关基因 Genes related to self-replication | rRNA基因 | rrn16a,rrn23a,rrn4.5a,rrn5a |
| tRNA基因 | trnA-UGCa,b,trnC-GCA,trnD-GUC,trnE-UUC,trnF-GAA,trnfM-CAU,trnG-GCCb,trnG-UCC,trnH-GUG,trnI-CAUa,trnI-GAUa,b,trnK-UUUb,trnL-CAAa,trnL-UAAb,trnL-UAG,trnM-CAU,trnN-GUUa,trnP-UGG,trnQ-UUG,trnR-ACGa,trnR-UCU,trnS-GCU,trnS-GGA,trnS-UGA,trnT-GGU,trnT-UGU,trnV-GAC,trnV-UACa,b,trnW-CCA,trnY-GUA | |
| 核糖体小亚基 | rps11,rps12a,c,rps14,rps15,rps16b,rps18,rps19,rps2 | |
| 核糖体大亚基 | rps3,rps4,rps7a,rps8,rpl14,rpl16b,rpl2a,b,rpl20,rpl22,rpl23a,rpl32,rpl33 | |
| DNA依赖的RNA聚合酶 | rpl36,rpoA,rpoB,rpoC1b,rpoC2 | |
| 其他功能基因 Other functional genes | 成熟酶 | matK |
| 蛋白质膜基因 | cemA | |
| 乙酰辅酶A亚基 | accD | |
| C类型细胞色素合成基因 | ccsA | |
| 蛋白酶 | clpPc | |
| 未知功能基因 Functionally unknown genes | 保守开放阅读框架 | ycf1a,ycf2a,ycf3c,ycf4 |
表2 海甘蓝叶绿体基因组基因功能总结
Table 2 Summary of assembled gene functions of C. abyssinica chloroplast genome
| 基因功能分类 Category for genes | 基因功能 Groups of genes | 基因名 Names of genes |
|---|---|---|
| 参与光合作用的基因 Genes involving in photosynthesis | 光合系统亚基 | ndhI,ndhJ,ndhK,psaA,psaB,psaC,psaI,psaJ,psbA,psbB,psbC,psbD,psbE,psbF,psbH,psbI,psbJ,psbK,psbL,psbM,psbN,psbT,psbZ |
| 细胞色素复合体亚基 | petA,petBb,petDb,petG,petL,petN | |
| 二磷酸核酮糖羧化酶大亚基 | rbcL | |
| ATP合成酶亚基 | atpA,atpB,atpE,atpFb,atpH,atpI | |
| NADH脱氢酶亚基 | ndhAb,ndhBa,b,ndhC,ndhD,ndhE,ndhF,ndhG,ndhH | |
| 自身复制相关基因 Genes related to self-replication | rRNA基因 | rrn16a,rrn23a,rrn4.5a,rrn5a |
| tRNA基因 | trnA-UGCa,b,trnC-GCA,trnD-GUC,trnE-UUC,trnF-GAA,trnfM-CAU,trnG-GCCb,trnG-UCC,trnH-GUG,trnI-CAUa,trnI-GAUa,b,trnK-UUUb,trnL-CAAa,trnL-UAAb,trnL-UAG,trnM-CAU,trnN-GUUa,trnP-UGG,trnQ-UUG,trnR-ACGa,trnR-UCU,trnS-GCU,trnS-GGA,trnS-UGA,trnT-GGU,trnT-UGU,trnV-GAC,trnV-UACa,b,trnW-CCA,trnY-GUA | |
| 核糖体小亚基 | rps11,rps12a,c,rps14,rps15,rps16b,rps18,rps19,rps2 | |
| 核糖体大亚基 | rps3,rps4,rps7a,rps8,rpl14,rpl16b,rpl2a,b,rpl20,rpl22,rpl23a,rpl32,rpl33 | |
| DNA依赖的RNA聚合酶 | rpl36,rpoA,rpoB,rpoC1b,rpoC2 | |
| 其他功能基因 Other functional genes | 成熟酶 | matK |
| 蛋白质膜基因 | cemA | |
| 乙酰辅酶A亚基 | accD | |
| C类型细胞色素合成基因 | ccsA | |
| 蛋白酶 | clpPc | |
| 未知功能基因 Functionally unknown genes | 保守开放阅读框架 | ycf1a,ycf2a,ycf3c,ycf4 |
| 基因组结构 Genome features | 海甘蓝 C. abyssinica | 拟南芥 A. thaliana | 克拉里克海甘蓝 C. kralikii | 甘蓝型油菜 B. napus | 萝卜 R. sativus | 白菜 B. rapa |
|---|---|---|---|---|---|---|
| 基因组大小Genome size/bp | 153 754 | 154 478 | 153 919 | 152 860 | 153 368 | 153 482 |
| 大单拷贝区LSC length/bp | 83 622 | 84 170 | 83 688 | 83 030 | 83 170 | 83 282 |
| 小单拷贝区SSC length/bp | 17 780 | 17 780 | 17 769 | 17 760 | 17 764 | 17 775 |
| 反向重复区IR length/bp | 26 176 | 26 264 | 26 231 | 26 035 | 26 217 | 26 213 |
| GC含量GC content/% | 36.37 | 36.29 | 36.35 | 36.32 | 36.34 | 36.36 |
| 基因数量Number of genes | 111 | 113 | 113 | 113 | 113 | 113 |
| 蛋白编码基因Protein-coding gene | 78 | 79 | 79 | 79 | 79 | 79 |
| rRNA | 4 | 4 | 4 | 4 | 4 | 4 |
| tRNA | 29 | 30 | 30 | 30 | 30 | 30 |
表3 6种十字花科物种叶绿体基因组特征比较
Table 3 Comparison of six chloroplast genomes of Brassicaceae species
| 基因组结构 Genome features | 海甘蓝 C. abyssinica | 拟南芥 A. thaliana | 克拉里克海甘蓝 C. kralikii | 甘蓝型油菜 B. napus | 萝卜 R. sativus | 白菜 B. rapa |
|---|---|---|---|---|---|---|
| 基因组大小Genome size/bp | 153 754 | 154 478 | 153 919 | 152 860 | 153 368 | 153 482 |
| 大单拷贝区LSC length/bp | 83 622 | 84 170 | 83 688 | 83 030 | 83 170 | 83 282 |
| 小单拷贝区SSC length/bp | 17 780 | 17 780 | 17 769 | 17 760 | 17 764 | 17 775 |
| 反向重复区IR length/bp | 26 176 | 26 264 | 26 231 | 26 035 | 26 217 | 26 213 |
| GC含量GC content/% | 36.37 | 36.29 | 36.35 | 36.32 | 36.34 | 36.36 |
| 基因数量Number of genes | 111 | 113 | 113 | 113 | 113 | 113 |
| 蛋白编码基因Protein-coding gene | 78 | 79 | 79 | 79 | 79 | 79 |
| rRNA | 4 | 4 | 4 | 4 | 4 | 4 |
| tRNA | 29 | 30 | 30 | 30 | 30 | 30 |
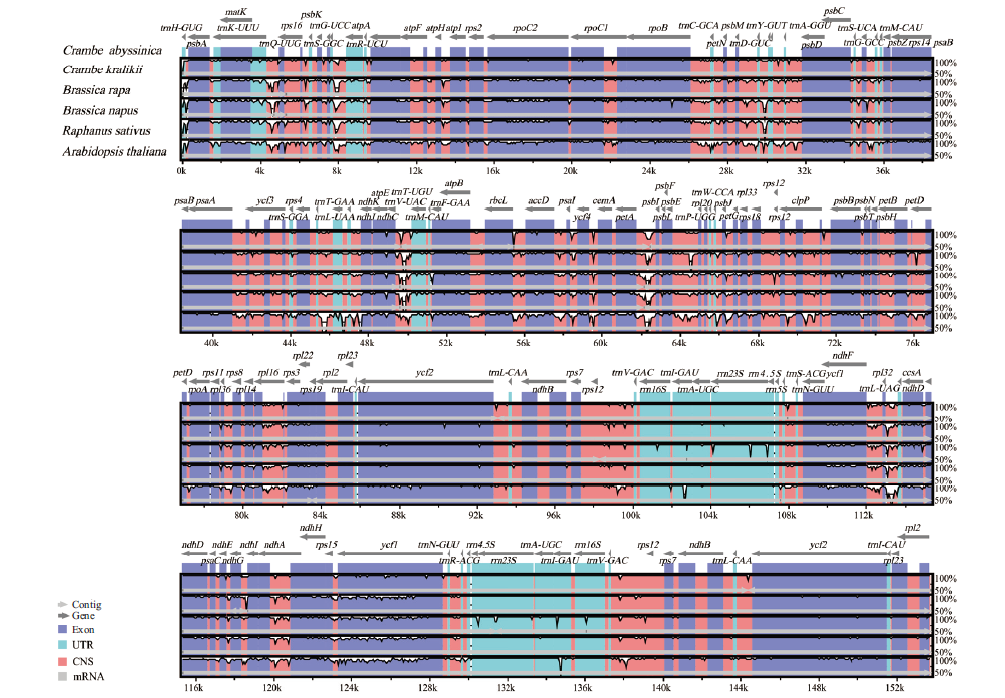
图2 以海甘蓝为参考与其他5个近缘物种的序列比对结果 灰色箭头和粗黑线表明基因定位。紫色条代表外显子,蓝色条代表非翻译区(UTR),粉色条代表非编码序列(CNS),灰色条代表mRNA,白色峰代表基因组学差异。y轴表示同一性百分比(显示:50%-100%)
Fig.2 Sequence alignment of 5 chloroplast genomes of Brassicaceae,with the annotation of C. abyssinica as reference Gray arrows above the alignment indicate the position and direction of each gene. The Y-scale represents the percent of identity between 50% and 100%. Genome regions are color coded as exon,tRNA,rRNA and conserved noncoding sequences(CNS)
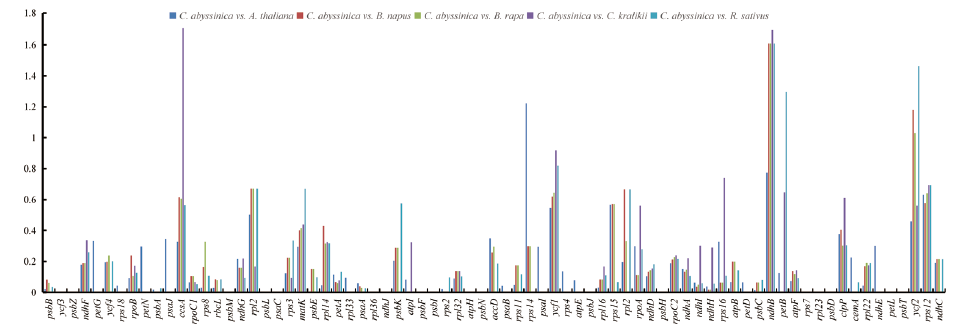
图5 海甘蓝与十字花科5个近缘物种叶绿体基因组中75个蛋白编码基因的Ka/Ks值
Fig.5 Ka/Ks ratios of 75 PCGs of the C. abyssinica chloroplast genome versus 5 closely related species of Brassicaceae
| [1] |
Francisco-Ortega J, Fuertes-Aguilar J, Gómez-Campo C, et al. Internal transcribed spacer sequence phylogeny of Crambe L. (Brassicaceae):molecular data reveal two old world disjunctions[J]. Mol Phylogenetics Evol, 1999, 11(3):361-380.
doi: 10.1006/mpev.1998.0592 URL |
| [2] | 王幼平, 罗鹏, 高宏波. 高芥酸油料植物海甘蓝的引种和利用[J]. 植物学通报, 1998, 33(3):23 |
| Wang YP, Luo P, Gao HB. Introduction and utilization of high erucic acid oil plant Crambe abyssinica[J]. Chin Bull Bot, 1998, 33(3):23 | |
| [3] | 胡金. 海甘蓝对重金属耐性与积累的研究[D]. 武汉: 湖北大学, 2016. |
| Hu J. Tolerance to and accumulation of heavy metals in Crambe abyssinica[D]. Wuhan: Hubei University, 2016. | |
| [4] | 王幼平. 高芥酸油料植物资源的研究[J]. 生物学通报, 1997, 32(12):41-43. |
| Wang YP. Study on resources of high erucic acid oil plant[J]. Bull Biol, 1997, 32(12):41-43. | |
| [5] |
Lazzeri L, de Mattei F, Bucelli F, et al. Crambe oil - a potential new hydraulic oil and quenchant[J]. Ind Lubr Tribol, 1997, 49(2):71-77.
doi: 10.1108/00368799710163893 URL |
| [6] | 王幼平, 罗鹏, 贾勇炯, 等. 海甘蓝与芸薹属属间杂交的受精和胚胎发育[J]. 作物学报, 1997, 23(5):538-544, 641-642. |
| Wang YP, Luo P, Jia YJ, et al. Fertilization and embryo development in generic crosses between Brassica species and C. abyssinica[J]. Acta Agron Sin, 1997, 23(5):538-544, 641-642. | |
| [7] |
Zhu B, Tu Y, Zeng P, et al. Extraction of the constituent subgenomes of the natural allopolyploid rapeseed(Brassica napus L.)[J]. Genetics, 2016, 204(3):1015-1027.
doi: 10.1534/genetics.116.190967 URL |
| [8] | 蔡梦鲜, 闫贵欣, 伍晓明, 等. 油菜叶绿体基因组研究进展[J]. 中国油料作物学报, 2013, 10:10-15. |
| Cai MX, Yan GX, Wu XM, et al. Advances in chloroplast genome of Brassica[J]. Chinese Journal of Oil Crop Sciences, 2013, 10:10-15. | |
| [9] |
Timmis JN, Ayliffe MA, Huang CY, et al. Endosymbiotic gene transfer:organelle genomes forge eukaryotic chromosomes[J]. Nat Rev Genet, 2004, 5(2):123-135.
doi: 10.1038/nrg1271 URL |
| [10] | 杨家鑫. 掌叶木和伞花木的叶绿体全基因组研究及其系统位置分析[D]. 贵阳: 贵州大学, 2019. |
| Yang JX. The complete chloroplast genome and phylogenetic position of Handeliodendron bodinieri and Eurycorymbus cavaleriei[D]. Guiyang: Guizhou University, 2019. | |
| [11] |
Branlant C, Krol A, Machatt MA, et al. Primary and secondary structures of Escherichia coli MRE 600 23S ribosomal RNA. Comparison with models of secondary structure for maize chloroplast 23S rRNA and for large portions of mouse and human 16S mitochondrial rRNAs[J]. Nucl Acids Res, 1981, 9(17):4303-4324.
doi: 10.1093/nar/9.17.4303 URL |
| [12] |
Palmer JD. Comparative organization of chloroplast genomes[J]. Annu Rev Genet, 1985, 19(1):325-354.
doi: 10.1146/annurev.ge.19.120185.001545 URL |
| [13] |
Wicke S, Schneeweiss GM, De Pamphilis CW, et al. The evolution of the plastid chromosome in land plants:gene content, gene order, gene function[J]. Plant Molecular Biology, 2011, 76(3-5):273-297.
doi: 10.1007/s11103-011-9762-4 URL |
| [14] |
Wolfe KH, Li WH, Sharp PM. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs[J]. PNAS, 1987, 84(24):9054-9058.
pmid: 3480529 |
| [15] |
Zhou JG, Chen XL, Cui YX, et al. Molecular structure and phylogenetic analyses of complete chloroplast genomes of two Aristolochia medicinal species[J]. Int J Mol Sci, 2017, 18(9):E1839.
doi: 10.3390/ijms18091839 URL |
| [16] |
Zhu B, Gao ZM, Luo X, et al. The complete chloroplast genome sequence of garden cress(Lepidium sativum L.)and its phylogenetic analysis in Brassicaceae family[J]. Mitochondrial DNA Part B, 2019, 4(2):3601-3602.
doi: 10.1080/23802359.2019.1677527 URL |
| [17] |
朱斌, 甘晨晨, 王洪程. 球花石斛(Dendrobium thyrsiflorum)叶绿体基因组特征及亲缘关系解析[J]. 生物技术通报, 2021, 37(5):38-47.
doi: 10.13560/j.cnki.biotech.bull.1985.2020-1380 |
| Zhu B, Gan CC, Wang HC. Characteristics of the complete chloroplast genome of Dendrobium thyrsiflorum and its phylogenetic relationship analysis[J]. Biotechnol Bull, 2021, 37(5):38-47. | |
| [18] |
Ohyama K, Fukuzawa H, Kohchi T, et al. Chloroplast gene organization deduced from complete sequence of liverwort Marchantia polymorpha chloroplast DNA[J]. Nature, 1986, 322(6079):572-574.
doi: 10.1038/322572a0 URL |
| [19] |
Shinozaki K, Ohme M, Tanaka M, et al. The complete nucleotide sequence of the tobacco chloroplast genome:its gene organization and expression[J]. EMBO J, 1986, 5(9):2043-2049.
doi: 10.1002/j.1460-2075.1986.tb04464.x pmid: 16453699 |
| [20] | Dierckxsens N, Mardulyn P, Smits G. NOVOPlasty:de novo assembly of organelle genomes from whole genome data[J]. Nucleic Acids Res, 2017, 45(4):e18. |
| [21] | Tillich M, Lehwark P, Pellizzer T, et al. GeSeq-versatile and accurate annotation of organelle genomes[J]. Nucleic Acids Res, 2017, 45(w1):W6-W11. |
| [22] | Lohse M, Drechsel O, Kahlau S, et al. OrganellarGenomeDRAW——a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets[J]. Nucleic Acids Res, 2013, 41(web server issue):W575-W581. |
| [23] |
Kurtz S, Choudhuri JV, Ohlebusch E, et al. REPuter:the manifold applications of repeat analysis on a genomic scale[J]. Nucleic Acids Res, 2001, 29(22):4633-4642.
pmid: 11713313 |
| [24] |
Beier S, Thiel T, Münch T, et al. MISA-web:a web server for microsatellite prediction[J]. Bioinformatics, 2017, 33(16):2583-2585.
doi: 10.1093/bioinformatics/btx198 URL |
| [25] | Frazer KA, Pachter L, Poliakov A, et al. VISTA:computational tools for comparative genomics[J]. Nucleic Acids Res, 2004, 32(web server issue):W273-W279. |
| [26] |
Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7:improvements in performance and usability[J]. Mol Biol Evol, 2013, 30(4):772-780.
doi: 10.1093/molbev/mst010 URL |
| [27] |
Wang D, Zhang Y, Zhang Z, et al. KaKs_Calculator 2. 0:a toolkit incorporating gamma-series methods and sliding window strategies[J]. Genomics Proteomics Bioinformatics, 2010, 8(1):77-80.
doi: 10.1016/S1672-0229(10)60008-3 URL |
| [28] |
Kumar S, Stecher G, Tamura K. MEGA7:molecular evolutionary genetics analysis version 7. 0 for bigger datasets[J]. Mol Biol Evol, 2016, 33(7):1870-1874.
doi: 10.1093/molbev/msw054 URL |
| [29] |
Soltis PS, Soltis DE. The role of genetic and genomic attributes in the success of polyploids[J]. PNAS, 2000, 97(13):7051-7057.
pmid: 10860970 |
| [30] | Palmer JD. Plastid chromosomes:structure and evolution[M]//The molecular biology of plastids. Amsterdam:Elsevier, 1991:5-53. |
| [31] |
Zhu A, Guo W, Gupta S, et al. Evolutionary dynamics of the plastid inverted repeat:the effects of expansion, contraction, and loss on substitution rates[J]. New Phytol, 2016, 209(4):1747-1756.
doi: 10.1111/nph.13743 URL |
| [32] |
Guo XY, Liu JQ, Hao GQ, et al. Plastome phylogeny and early diversification of Brassicaceae[J]. BMC Genom, 2017, 18(1):176.
doi: 10.1186/s12864-017-3555-3 URL |
| [33] | Seol YJ, Kim K, Kang SH, et al. The complete chloroplast genome of two Brassica species, Brassica nigra and B. Oleracea[J]. Mitochondrial DNA A DNA Mapp Seq Anal, 2017, 28(2):167-168. |
| [34] | 包维红. 基于叶绿体基因组的猕猴桃科分子系统学研究[D]. 武汉: 中国科学院大学(中国科学院武汉植物园), 2018. |
| Bao WH. A phylogenetic study of Actinidiaceae based on chloroplast genomes[D]. Wuhan: University of Chinese Academy of Sciences(The Wuhan Botanical Garden of the Chinese Academy of Sciences), 2018. | |
| [35] | 沈立群. 唇形科三种药用植物叶绿体全基因组及科内的比较与进化分析[D]. 杭州: 浙江大学, 2018. |
| Shen LQ. Complete chloroplast genomes of three Lamiaceae medicinal plants:comparative and evolutionary analysis in Lamiaceae[D]. Hangzhou: Zhejiang University, 2018. | |
| [36] | Zhu B, Qian F, Hou Y, et al. Complete chloroplast genome features and phylogenetic analysis of Eruca sativa(Brassicaceae)[J]. PLoS One, 2021, 16(3):e0248556. |
| [37] | Du X, Zeng T, Feng Q, et al. The complete chloroplast genome sequence of yellow mustard(Sinapis alba L.)and its phylogenetic relationship to other Brassicaceae species[J]. Gene, 2020, 731:144340. |
| [38] |
Kim KJ, Lee HL. Complete chloroplast genome sequences from Korean ginseng(Panax schinseng Nees)and comparative analysis of sequence evolution among 17 vascular plants[J]. DNA Res, 2004, 11(4):247-261.
pmid: 15500250 |
| [39] |
Terakami S, Matsumura Y, Kurita K, et al. Complete sequence of the chloroplast genome from pear(Pyrus pyrifolia):genome structure and comparative analysis[J]. Tree Genet Genomes, 2012, 8(4):841-854.
doi: 10.1007/s11295-012-0469-8 URL |
| [40] | Qian J, Song J, Gao H, et al. The complete chloroplast genome sequence of the medicinal plant Salvia miltiorrhiza[J]. PLoS One, 2013, 8(2):e57607. |
| [41] | Zong D, Zhou A, Zhang Y, et al. Characterization of the complete chloroplast genomes of five Populus species from the western Sichuan plateau, southwest China:comparative and phylogenetic analyses[J]. PeerJ, 2019, 7:e6386. |
| [42] | Li Y, Zhang Z, Yang J, et al. Complete chloroplast genome of seven Fritillaria species, variable DNA markers identification and phylogenetic relationships within the genus[J]. PLoS One, 2018, 13(3):e0194613. |
| [43] | Cavalier-Smith T. Chloroplast evolution:secondary symbiogenesis and multiple losses[J]. Curr Biol, 2002, 12(2):R62-R64. |
| [44] | Nie X, Lv S, Zhang Y, et al. Complete chloroplast genome sequence of a major invasive species, crofton weed(Ageratina adenophora)[J]. PLoS One, 2012, 7(5):e36869. |
| [45] |
Powell W, Morgante M, McDevitt R, et al. Polymorphic simple sequence repeat regions in chloroplast genomes:applications to the population genetics of pines[J]. PNAS, 1995, 92(17):7759-7763.
pmid: 7644491 |
| [46] |
Pugh T, Fouet O, Risterucci AM, et al. A new cacao linkage map based on codominant markers:development and integration of 201 new microsatellite markers[J]. Theor Appl Genet, 2004, 108(6):1151-1161.
pmid: 14760486 |
| [47] |
Yan C, Du JC, Gao L, et al. The complete chloroplast genome sequence of watercress(Nasturtium officinale R. Br. ):Genome organization, adaptive evolution and phylogenetic relationships in Cardamineae[J]. Gene, 2019, 699:24-36.
doi: 10.1016/j.gene.2019.02.075 URL |
| [48] |
Kuang DY, Wu H, Wang YL, et al. Complete chloroplast genome sequence of Magnolia kwangsiensis(Magnoliaceae):implication for DNA barcoding and population genetics[J]. Genome, 2011, 54(8):663-673.
doi: 10.1139/g11-026 URL |
| [49] | Asaf S, Waqas M, Khan AL, et al. The complete chloroplast genome of wild rice(Oryza minuta)and its comparison to related species[J]. Front Plant Sci, 2017, 8:304. |
| [50] |
Morton BR. Strand asymmetry and codon usage bias in the chloroplast genome of Euglena gracilis[J]. PNAS, 1999, 96(9):5123-5128.
pmid: 10220429 |
| [51] |
Sharp PM, Li WH. The codon adaptation index——a measure of directional synonymous codon usage bias, and its potential applications[J]. Nucleic Acids Res, 1987, 15(3):1281-1295.
pmid: 3547335 |
| [52] | Mazumder TH, Chakraborty S. Gaining insights into the codon usage patterns of TP53 gene across eight mammalian species[J]. PLoS One, 2015, 10(3):e0121709. |
| [53] |
Jeong YM, Chung WH, Mun JH, et al. De novo assembly and characterization of the complete chloroplast genome of radish(Raphanus sativus L.)[J]. Gene, 2014, 551(1):39-48.
doi: 10.1016/j.gene.2014.08.038 URL |
| [54] |
Makalowski W, Boguski MS. Evolutionary parameters of the transcribed mammalian genome:an analysis of 2, 820 orthologous rodent and human sequences[J]. PNAS, 1998, 95(16):9407-9412.
pmid: 9689093 |
| [55] |
马艳莉, 陈海燕, 王继刚, 等. 银杏叶绿体ndhF RNA编辑现象分析及C290位编辑对胁迫处理的响应[J]. 植物学报, 2011, 46(1):1-10.
doi: 10.3724/SP.J.1259.2011.00001 |
| Ma YL, Chen HY, Wang JG, et al. Analysis of editing sites for chloroplast ndhF in Ginkgo biloba and the editing efficiency at C290 in response to different stresses[J]. Chin Bull Bot, 2011, 46(1):1-10. | |
| [56] |
Drescher A, Ruf S, Calsa T, et al. The two largest chloroplast genome-encoded open reading frames of higher plants are essential genes[J]. Plant J, 2000, 22(2):97-104.
pmid: 10792825 |
| [57] |
Wang WB, Yu H, Wang JH, et al. The complete chloroplast genome sequences of the medicinal plant Forsythia suspensa(Oleaceae)[J]. Int J Mol Sci, 2017, 18(11):2288.
doi: 10.3390/ijms18112288 URL |
| [58] | 朱斌, 胡利娟, 田忠静, 等. 基于叶绿体基因组序列对50种菊科植物亲缘关系分析[J]. 贵州师范大学学报:自然科学版, 2021, 39(2):15-25. |
| Zhu B, Hu LJ, Tian ZJ, et al. Phylogenetic relationship analysis of 50 species in Asteraceae family based on the chloroplast genome sequence[J]. J Guizhou Norm Univ:Nat Sci, 2021, 39(2):15-25. | |
| [59] |
Warwick SI, Sauder CA. Phylogeny of tribe Brassiceae(Brassicaceae)based on chloroplast restriction site polymorphisms and nuclear ribosomal internal transcribed spacer and chloroplast trnL intron sequences[J]. Can J Bot, 2005, 83(5):467-483.
doi: 10.1139/b05-021 URL |
| [60] | 程雨贵, 吴江生, 陈洪高, 等. 萝卜和甘蓝远缘杂交研究[J]. 中国油料作物学报, 2006, 28(1):1-6. |
| Cheng YG, Wu JS, Chen HG, et al. Studies on intergeneric hybridization between Raphanus sativus and Brassica olerace[J]. Chin J Oil Crop Sci, 2006, 28(1):1-6. |
| [1] | 尹明华, 余锾媛, 肖心怡, 王玉婷. 江西铅山红芽芋叶绿体基因组特征及系统发育分析[J]. 生物技术通报, 2023, 39(6): 233-247. |
| [2] | 曲春娟, 朱悦, 江晨, 曲明静, 王向誉, 李晓. 铜绿丽金龟线粒体全基因组及其系统发育分析[J]. 生物技术通报, 2023, 39(2): 263-273. |
| [3] | 刘雄伟, 刘畅, 曾宪法, 杨小英, 俸婷婷, 赵杰宏, 周英. 朱砂根叶绿体全基因组解析及系统发育分析[J]. 生物技术通报, 2023, 39(1): 232-242. |
| [4] | 刘警鞠, 张雨森, 陈娟, 孙炳达, 赵国柱. 曲霉属的现代分类命名研究进展[J]. 生物技术通报, 2022, 38(7): 109-118. |
| [5] | 朱斌, 甘晨晨, 王洪程. 球花石斛(Dendrobium thyrsiflorum)叶绿体基因组特征及亲缘关系解析[J]. 生物技术通报, 2021, 37(5): 38-47. |
| [6] | 程赛赛, 龚鑫, 薛文凤, 万兵兵, 刘满强, 胡锋. 生态进化视角的植物-微生物组互作研究进展[J]. 生物技术通报, 2020, 36(9): 3-13. |
| [7] | 李裕华, 任永康, 赵兴华, 刘江, 韩斌, 王长彪, 唐朝晖. 禾本科主要农作物叶绿体基因组研究进展[J]. 生物技术通报, 2020, 36(11): 112-121. |
| [8] | 刘凌燕, 陈志宇, 曾还雄, 林培彬, 金小宝. 美洲大蠊肠道内生微杆菌的分离鉴定及其抑菌活性研究[J]. 生物技术通报, 2018, 34(6): 172-177. |
| [9] | 张广志, 王加宁, 吴晓青, 周方园, 张新建, 赵晓燕, 谢雪迎, 周红姿. 设施番茄根围土样中木霉菌多样性及功能活性分析[J]. 生物技术通报, 2018, 34(4): 179-185. |
| [10] | 胡文哲, 谭泽文, 王勇, 徐羡微, 谭志远. 藤县药用野生稻内生固氮菌分离鉴定及系统发育分析[J]. 生物技术通报, 2016, 32(6): 111-119. |
| [11] | 李丽卡, 李象钦, 谢国文, 李海生, 郑毅胜, 藤婕华, 高锦伟. 基于cpDNA片段探讨中-日间断分布双花木属植物的系统发育学[J]. 生物技术通报, 2016, 32(1): 80-87. |
| [12] | 刘国红, 刘波, 林营志, 唐建阳. 基于脂肪酸生物标记与16S rRNA的芽胞杆菌系统发育分析比较[J]. 生物技术通报, 2015, 31(3): 146-153. |
| [13] | 罗静初. 实用生物信息技术课程教学实例[J]. 生物技术通报, 2015, 31(11): 102-111. |
| [14] | 焦林培金,黄运红,李素珍,龙中儿. 一株拮抗链霉菌的鉴定及其多相分类特征[J]. 生物技术通报, 2015, 31(1): 138-143. |
| [15] | 陈秀玲, 李景富, 张丽莉, 张俊峰, 王傲雪. 一株淡色生赤壳菌的生防作用分析及系统发育树构建[J]. 生物技术通报, 2014, 0(5): 184-189. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||