








生物技术通报 ›› 2023, Vol. 39 ›› Issue (3): 89-100.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0810
余世洲1( ), 曹领改1, 王世泽1,2, 刘勇1,3, 边文杰4, 任学良1(
), 曹领改1, 王世泽1,2, 刘勇1,3, 边文杰4, 任学良1( )
)
收稿日期:2022-06-30
出版日期:2023-03-26
发布日期:2023-04-10
通讯作者:
任学良,男,博士,研究员,研究方向:烟草遗传育种;E-mail:renxl@126.com作者简介:余世洲,男,博士,副研究员,研究方向:烟草遗传育种;E-mail:yusz@nwafu.edu.cn
基金资助:
YU Shi-zhou1( ), CAO Ling-gai1, WANG Shi-ze1,2, LIU Yong1,3, BIAN Wen-jie4, REN Xue-liang1(
), CAO Ling-gai1, WANG Shi-ze1,2, LIU Yong1,3, BIAN Wen-jie4, REN Xue-liang1( )
)
Received:2022-06-30
Published:2023-03-26
Online:2023-04-10
摘要:
基于KASP(kompetitive allele specific PCR)技术平台,开发并验证可用于烟草核心种质基因分型的SNP(single nucleotide polymorphism)标记和对应检测引物,为烟草种质基因型鉴定评价、遗传多样性分析、核心种质筛选等提供技术和数据支持。利用Python和Perl脚本程序对覆盖烟草全基因组的1 179 154个SNP位点进行KASP引物设计和筛选,并通过试验验证其准确度和可用性。结果共有217 621个SNP位点完成了对应KASP引物设计,选择1 378个SNP位点进行试验验证,明确了732个可作为SNP标记,并确定48个SNP标记作为烟草种质资源基因分型的核心标记。这48个核心标记在烟草24条染色体上平均分布,平均PIC为0.36,平均MAF为0.39。利用确定的48个核心SNP标记,可以将各供试种质特别是将当前主栽烟草品种基因型进行区分,且标记具有极高的可靠性。
余世洲, 曹领改, 王世泽, 刘勇, 边文杰, 任学良. 烟草种质基因分型核心SNP标记的开发[J]. 生物技术通报, 2023, 39(3): 89-100.
YU Shi-zhou, CAO Ling-gai, WANG Shi-ze, LIU Yong, BIAN Wen-jie, REN Xue-liang. Development Core SNP Markers for Tobacco Germplasm Genotyping[J]. Biotechnology Bulletin, 2023, 39(3): 89-100.
| 序号 No. | 种质库编号 GenBank number | 种质名称 Accession name | 调制类型 Curing status | 序号 No. | 资源库编号 GenBank number | 种质名称 Accession name | 调制类型 Curing status | |
|---|---|---|---|---|---|---|---|---|
| 1 | FD002 | 独山软杆Dushanruangan | 烤烟 Flue-cured | 76 | XZ018 | PX5 | 烤烟 Flue-cured | |
| 2 | FD011 | 福泉团叶摺烟 Fuquan tanyezheyan | 烤烟Flue-cured | 77 | XZ019 | PX6 | 烤烟 Flue-cured | |
| 3 | FD032 | 麻将大红花 Majiangdahonghua | 烤烟Flue-cured | 78 | XZ020 | PX7 | 烤烟 Flue-cured | |
| 4 | FD041 | 黔江一号Qianjiangyihao | 烤烟Flue-cured | 79 | XZ021 | PX8 | 烤烟 Flue-cured | |
| 5 | FD047 | 瓮安中坪烤烟 Wenganzhongpingkaoyan | 烤烟 Flue-cured | 80 | XZ022 | PX9 | 烤烟 Flue-cured | |
| 6 | FS054 | 柳叶尖0695 Liuyejian 0695 | 烤烟 Flue-cured | 81 | XZ023 | PX10 | 烤烟 Flue-cured | |
| 7 | FS070 | 胎里富1011 Tailifu 1011 | 烤烟 Flue-cured | 82 | XZ024 | PX11 | 烤烟 Flue-cured | |
| 8 | FS071 | 歪把子Waibazi | 烤烟 Flue-cured | 83 | XZ025 | PX12 | 烤烟 Flue-cured | |
| 9 | FY064 | 哲伍毛烟Zhewumaoyan | 烤烟 Flue-cured | 84 | XZ026 | PX13 | 烤烟 Flue-cured | |
| 10 | FY061 | 片片黄Pianpianhuang | 烤烟 Flue-cured | 85 | XZ027 | PX14 | 烤烟 Flue-cured | |
| 11 | FG015 | Coker86 | 烤烟 Flue-cured | 86 | XZ028 | PX15 | 烤烟 Flue-cured | |
| 12 | FG018 | Dixie Bright 101 | 烤烟 Flue-cured | 87 | XZ029 | PX17 | 烤烟 Flue-cured | |
| 13 | FG024 | Hicks | 烤烟 Flue-cured | 88 | XZ030 | PX19 | 烤烟 Flue-cured | |
| 14 | FG027 | K149 | 烤烟 Flue-cured | 89 | SG003 | 安龙护耳大柳叶 Anlonghuerdaliuye | 晒烟 Sun-cured | |
| 15 | FG029 | K346 | 烤烟 Flue-cured | 90 | SG009 | 安顺二吊枝 Anshunerdiaozhi | 晒烟 Sun-cured | |
| 16 | FG028 | K326 | 烤烟 Flue-cured | 91 | SG048 | 大方大红花 Dafangdahonghua | 晒烟 Sun-cured | |
| 17 | FG047 | NC567 | 烤烟 Flue-cured | 92 | SG052 | 大柳叶节骨密 Daliuyegumi | 晒烟 Sun-cured | |
| 18 | FG049 | NC729 | 烤烟 Flue-cured | 93 | SG069 | 德江中花烟(2) Dejiangzhonghuayan(2) | 晒烟 Sun-cured | |
| 19 | FG052 | NC82 | 烤烟 Flue-cured | 94 | SG079 | 光柄柳叶(罗) Guangbingliuye(luo) | 晒烟 Sun-cured | |
| 20 | FG053 | NC89 | 烤烟 Flue-cured | 95 | SG080 | 光柄柳叶(木) Guangbingliuye(mu) | 晒烟 Sun-cured | |
| 21 | FG054 | NC95 | 烤烟 Flue-cured | 96 | SG094 | 花溪大青杆 Huaxidaqinggan | 晒烟 Sun-cured | |
| 22 | FG061 | 牛津26 Oxford 26 | 烤烟 Flue-cured | 97 | SG102 | 惠水三都大白花 Huishuisandudabaihua | 晒烟 Sun-cured | |
| 23 | FG077 | RG11 | 烤烟 Flue-cured | 98 | SG112 | 凯里枇杷烟 Kailipipayan | 晒烟 Sun-cured | |
| 24 | FG079 | RG17 | 烤烟 Flue-cured | 99 | SG134 | 马耳烟Maeryan | 晒烟 Sun-cured | |
| 25 | FG080 | RG8 | 烤烟 Flue-cured | 100 | SG156 | 盘县大柳叶Panxiandaliuye | 晒烟 Sun-cured | |
| 26 | FG089 | Special 400 | 烤烟 Flue-cured | 101 | SG169 | 平塘中坝晒烟 Pingtangzhongbashaiyan | 晒烟 Sun-cured | |
| 27 | FG090 | Special 401 | 烤烟 Flue-cured | 102 | SG170 | 普安寸骨烟 Puancunguyan | 晒烟 Sun-cured | |
| 28 | FG094 | SpeightG-164 | 烤烟 Flue-cured | 103 | SG178 | 黔西红花吊把 Qianxihonghuadiaoba | 晒烟 Sun-cured | |
| 29 | FG095 | Speight G-28 | 烤烟 Flue-cured | 104 | SG248 | 贞丰柳叶烟 Zhenfengliuyeyan | 晒烟 Sun-cured | |
| 30 | FG099 | Speight G-80 | 烤烟 Flue-cured | 105 | SG258 | 镇远莲花烟(周) Zhenyuanlianhuayan(zhou) | 晒烟 Sun-cured | |
| 31 | FG100 | SPTG-172 | 烤烟 Flue-cured | 106 | SS006 | 81—26 | 晒烟 Sun-cured | |
| 32 | FG105 | Ti448A | 烤烟 Flue-cured | 107 | SS012 | Va309 | 晒烟 Sun-cured | |
| 33 | FG118 | Vesta 47 | 烤烟 Flue-cured | 108 | SS021 | 矮株一号Aizhuyihao | 晒烟 Sun-cured | |
| 34 | FG120 | 佛光Foguang | 烤烟 Flue-cured | 109 | SS027 | 把柄烟Babingyan | 晒烟 Sun-cured | |
| 35 | FG122 | Yellow mammoth | 烤烟 Flue-cured | 110 | SS045 | 大瓦垅Dawalong | 晒烟 Sun-cured | |
| 36 | FG071 | PVH06 | 烤烟 Flue-cured | 111 | SS053 | 风林一号Fenglinyihao | 晒烟 Sun-cured | |
| 37 | XZ001 | CV87 | 烤烟 Flue-cured | 112 | SS066 | 贺县公会晒烟 Hexiangonghuishaiyan | 晒烟 Sun-cured | |
| 38 | XZ002 | Cash | 烤烟 Flue-cured | 113 | SS075 | 护脖香1368 Huboxiang 1368 | 晒烟 Sun-cured | |
| 39 | XZ003 | Coker139 | 烤烟 Flue-cured | 114 | SS087 | 老山烟Laoshanyan | 晒烟 Sun-cured | |
| 40 | XZ004 | Gold doller | 烤烟 Flue-cured | 115 | SS091 | 马里村晒烟(二) Malicunshaiyan(2) | 晒烟 Sun-cured | |
| 41 | XZ005 | Mcnair 30 | 烤烟 Flue-cured | 116 | SS097 | 勐板晒烟 Mengbanshaiyan | 晒烟 Sun-cured | |
| 42 | XZ006 | Orinoco | 烤烟 Flue-cured | 117 | SS113 | 千层塔Qiancengta | 晒烟 Sun-cured | |
| 43 | XZ007 | Mammoth gold | 烤烟 Flue-cured | 118 | SS132 | 腾冲大柳叶 Tengchongdaliuye | 晒烟 Sun-cured | |
| 44 | XZ008 | Golden wilf | 烤烟 Flue-cured | 119 | SS142 | 小叶尖Xiaoyejian | 晒烟 Sun-cured | |
| 45 | XZ009 | CV91 | 烤烟 Flue-cured | 120 | SS146 | 一朵花Yiduohua | 晒烟 Sun-cured | |
| 46 | XZ010 | 云烟315Yunyan 315 | 烤烟 Flue-cured | 121 | SS164 | 大枇杷烟Dapipayan | 晒烟 Sun-cured | |
| 47 | FS027 | 大黄金5210 Dahuangjin 5210 | 烤烟 Flue-cured | 122 | SS169 | 金山乡旱烟 Jinshanxianghanyan | 晒烟 Sun-cured | |
| 48 | FS029 | 单育2号Danyu No.2 | 烤烟 Flue-cured | 123 | XZ031 | K17 | 晒烟 Sun-cured | |
| 49 | FS037 | 广黄55 Guanghuang 55 | 烤烟 Flue-cured | 124 | XZ032 | 寸三皮Cunsanpi | 晒烟 Sun-cured | |
| 50 | FS044 | 红星一号Hongxingyihao | 烤烟 Flue-cured | 125 | XZ033 | 九月寒Jiuyuehan | 晒烟 Sun-cured | |
| 51 | FS045 | 金星6007 Jinxing 6007 | 烤烟 Flue-cured | 126 | XZ034 | 三生烟Sanshengyan | 晒烟 Sun-cured | |
| 52 | FS050 | 净叶黄Jingyehuang | 烤烟 Flue-cured | 127 | BG012 | W.B68 | 白肋烟 Burley | |
| 53 | FS061 | 潘园黄Panyuanhuang | 烤烟 Flue-cured | 128 | BG018 | ky41A | 白肋烟Burley | |
| 54 | FS077 | 许金2号Xujin No. 2 | 烤烟 Flue-cured | 129 | BG023 | 半铁泡Bantiepao | 白肋烟Burley | |
| 55 | FS084 | 云烟87 Yunyan 87 | 烤烟 Flue-cured | 130 | BG029 | BY21 | 白肋烟Burley | |
| 56 | FS085 | 云烟2号Yunyan No.2 | 烤烟 Flue-cured | 131 | BG032 | KY10 | 白肋烟Burley | |
| 57 | FS089 | 中烟90 Zhongyan 90 | 烤烟 Flue-cured | 132 | BG033 | KY14 | 白肋烟Burley | |
| 58 | FY017 | 春雷三号 Chunlei No.3 | 烤烟 Flue-cured | 133 | AG002 | 从江塘洞烟 Congjiangtangdongyan | 晾烟Air-cured | |
| 59 | FY030 | 湄潭黑团壳 Meitanheituanke | 烤烟 Flue-cured | 134 | AG018 | 台江小柳叶 Taijiangxiaoliuye | 晾烟Air-cured | |
| 60 | FS083 | 云烟85 Yunyan 85 | 烤烟 Flue-cured | 135 | AG006 | 剑河乡洞土烟 Jianhexiangdongtuyan | 晾烟Air-cured | |
| 61 | XZ011 | 云烟97 Yunyan 97 | 烤烟 Flue-cured | 136 | GG001 | Ambale-ma | 雪茄烟 Cigar | |
| 62 | FY048 | 兴烟一号Xingyanyihao | 烤烟 Flue-cured | 137 | GG002 | Beinhart1000-1 | 雪茄烟 Cigar | |
| 63 | FT011 | 南江3号Nanjiang No.3 | 烤烟 Flue-cured | 138 | GG004 | Florida301 | 雪茄烟Cigar | |
| 64 | FY068 | 遵烟6号Zunyan No.6 | 烤烟 Flue-cured | 139 | GG005 | Pennbel 69 | 雪茄烟Cigar | |
| 65 | XZ012 | 中烟100 Zhongyan 100 | 烤烟 Flue-cured | 140 | XZ035 | 包皮Baopi | 雪茄烟Cigar | |
| 66 | FS032 | 贵烟3号Guiyan No.3 | 烤烟 Flue-cured | 141 | XZ036 | 坦桑尼亚烟 Tansangniyayan | 香料烟 Oriental | |
| 67 | FS087 | 中烟14 Zhongyan 14 | 烤烟 Flue-cured | 142 | TG002 | Adcook | 香料烟Oriental | |
| 68 | FS032 | 革新3号Gexin No.3 | 烤烟 Flue-cured | 143 | TG004 | Basma536 | 香料烟Oriental | |
| 69 | PG142 | K326LF | 烤烟 Flue-cured | 144 | TG006 | Cekpka | 香料烟Oriental | |
| 70 | FY094 | Q6 | 烤烟 Flue-cured | 145 | TG011 | xanthi-nc | 香料烟Oriental | |
| 71 | XZ013 | 瓮安2号Wengan No.2 | 烤烟 Flue-cured | 146 | XZ037 | YY1 | 药烟 Medicinal | |
| 72 | XZ014 | 2010A4 | 烤烟 Flue-cured | 147 | XZ038 | YY2 | 药烟Medicinal | |
| 73 | XZ015 | 兴烟2号Xingyan No.2 | 烤烟 Flue-cured | 148 | XZ039 | YY4 | 药烟Medicinal | |
| 74 | XZ016 | 99215-1 | 烤烟 Flue-cured | 149 | XZ040 | YY11 | 药烟Medicinal | |
| 75 | XZ017 | PX2 | 烤烟 Flue-cured | 150 | XZ041 | YY15 | 药烟Medicinal |
表1 烟草种质资源列表
Table 1 List of tobacco germplasm resources
| 序号 No. | 种质库编号 GenBank number | 种质名称 Accession name | 调制类型 Curing status | 序号 No. | 资源库编号 GenBank number | 种质名称 Accession name | 调制类型 Curing status | |
|---|---|---|---|---|---|---|---|---|
| 1 | FD002 | 独山软杆Dushanruangan | 烤烟 Flue-cured | 76 | XZ018 | PX5 | 烤烟 Flue-cured | |
| 2 | FD011 | 福泉团叶摺烟 Fuquan tanyezheyan | 烤烟Flue-cured | 77 | XZ019 | PX6 | 烤烟 Flue-cured | |
| 3 | FD032 | 麻将大红花 Majiangdahonghua | 烤烟Flue-cured | 78 | XZ020 | PX7 | 烤烟 Flue-cured | |
| 4 | FD041 | 黔江一号Qianjiangyihao | 烤烟Flue-cured | 79 | XZ021 | PX8 | 烤烟 Flue-cured | |
| 5 | FD047 | 瓮安中坪烤烟 Wenganzhongpingkaoyan | 烤烟 Flue-cured | 80 | XZ022 | PX9 | 烤烟 Flue-cured | |
| 6 | FS054 | 柳叶尖0695 Liuyejian 0695 | 烤烟 Flue-cured | 81 | XZ023 | PX10 | 烤烟 Flue-cured | |
| 7 | FS070 | 胎里富1011 Tailifu 1011 | 烤烟 Flue-cured | 82 | XZ024 | PX11 | 烤烟 Flue-cured | |
| 8 | FS071 | 歪把子Waibazi | 烤烟 Flue-cured | 83 | XZ025 | PX12 | 烤烟 Flue-cured | |
| 9 | FY064 | 哲伍毛烟Zhewumaoyan | 烤烟 Flue-cured | 84 | XZ026 | PX13 | 烤烟 Flue-cured | |
| 10 | FY061 | 片片黄Pianpianhuang | 烤烟 Flue-cured | 85 | XZ027 | PX14 | 烤烟 Flue-cured | |
| 11 | FG015 | Coker86 | 烤烟 Flue-cured | 86 | XZ028 | PX15 | 烤烟 Flue-cured | |
| 12 | FG018 | Dixie Bright 101 | 烤烟 Flue-cured | 87 | XZ029 | PX17 | 烤烟 Flue-cured | |
| 13 | FG024 | Hicks | 烤烟 Flue-cured | 88 | XZ030 | PX19 | 烤烟 Flue-cured | |
| 14 | FG027 | K149 | 烤烟 Flue-cured | 89 | SG003 | 安龙护耳大柳叶 Anlonghuerdaliuye | 晒烟 Sun-cured | |
| 15 | FG029 | K346 | 烤烟 Flue-cured | 90 | SG009 | 安顺二吊枝 Anshunerdiaozhi | 晒烟 Sun-cured | |
| 16 | FG028 | K326 | 烤烟 Flue-cured | 91 | SG048 | 大方大红花 Dafangdahonghua | 晒烟 Sun-cured | |
| 17 | FG047 | NC567 | 烤烟 Flue-cured | 92 | SG052 | 大柳叶节骨密 Daliuyegumi | 晒烟 Sun-cured | |
| 18 | FG049 | NC729 | 烤烟 Flue-cured | 93 | SG069 | 德江中花烟(2) Dejiangzhonghuayan(2) | 晒烟 Sun-cured | |
| 19 | FG052 | NC82 | 烤烟 Flue-cured | 94 | SG079 | 光柄柳叶(罗) Guangbingliuye(luo) | 晒烟 Sun-cured | |
| 20 | FG053 | NC89 | 烤烟 Flue-cured | 95 | SG080 | 光柄柳叶(木) Guangbingliuye(mu) | 晒烟 Sun-cured | |
| 21 | FG054 | NC95 | 烤烟 Flue-cured | 96 | SG094 | 花溪大青杆 Huaxidaqinggan | 晒烟 Sun-cured | |
| 22 | FG061 | 牛津26 Oxford 26 | 烤烟 Flue-cured | 97 | SG102 | 惠水三都大白花 Huishuisandudabaihua | 晒烟 Sun-cured | |
| 23 | FG077 | RG11 | 烤烟 Flue-cured | 98 | SG112 | 凯里枇杷烟 Kailipipayan | 晒烟 Sun-cured | |
| 24 | FG079 | RG17 | 烤烟 Flue-cured | 99 | SG134 | 马耳烟Maeryan | 晒烟 Sun-cured | |
| 25 | FG080 | RG8 | 烤烟 Flue-cured | 100 | SG156 | 盘县大柳叶Panxiandaliuye | 晒烟 Sun-cured | |
| 26 | FG089 | Special 400 | 烤烟 Flue-cured | 101 | SG169 | 平塘中坝晒烟 Pingtangzhongbashaiyan | 晒烟 Sun-cured | |
| 27 | FG090 | Special 401 | 烤烟 Flue-cured | 102 | SG170 | 普安寸骨烟 Puancunguyan | 晒烟 Sun-cured | |
| 28 | FG094 | SpeightG-164 | 烤烟 Flue-cured | 103 | SG178 | 黔西红花吊把 Qianxihonghuadiaoba | 晒烟 Sun-cured | |
| 29 | FG095 | Speight G-28 | 烤烟 Flue-cured | 104 | SG248 | 贞丰柳叶烟 Zhenfengliuyeyan | 晒烟 Sun-cured | |
| 30 | FG099 | Speight G-80 | 烤烟 Flue-cured | 105 | SG258 | 镇远莲花烟(周) Zhenyuanlianhuayan(zhou) | 晒烟 Sun-cured | |
| 31 | FG100 | SPTG-172 | 烤烟 Flue-cured | 106 | SS006 | 81—26 | 晒烟 Sun-cured | |
| 32 | FG105 | Ti448A | 烤烟 Flue-cured | 107 | SS012 | Va309 | 晒烟 Sun-cured | |
| 33 | FG118 | Vesta 47 | 烤烟 Flue-cured | 108 | SS021 | 矮株一号Aizhuyihao | 晒烟 Sun-cured | |
| 34 | FG120 | 佛光Foguang | 烤烟 Flue-cured | 109 | SS027 | 把柄烟Babingyan | 晒烟 Sun-cured | |
| 35 | FG122 | Yellow mammoth | 烤烟 Flue-cured | 110 | SS045 | 大瓦垅Dawalong | 晒烟 Sun-cured | |
| 36 | FG071 | PVH06 | 烤烟 Flue-cured | 111 | SS053 | 风林一号Fenglinyihao | 晒烟 Sun-cured | |
| 37 | XZ001 | CV87 | 烤烟 Flue-cured | 112 | SS066 | 贺县公会晒烟 Hexiangonghuishaiyan | 晒烟 Sun-cured | |
| 38 | XZ002 | Cash | 烤烟 Flue-cured | 113 | SS075 | 护脖香1368 Huboxiang 1368 | 晒烟 Sun-cured | |
| 39 | XZ003 | Coker139 | 烤烟 Flue-cured | 114 | SS087 | 老山烟Laoshanyan | 晒烟 Sun-cured | |
| 40 | XZ004 | Gold doller | 烤烟 Flue-cured | 115 | SS091 | 马里村晒烟(二) Malicunshaiyan(2) | 晒烟 Sun-cured | |
| 41 | XZ005 | Mcnair 30 | 烤烟 Flue-cured | 116 | SS097 | 勐板晒烟 Mengbanshaiyan | 晒烟 Sun-cured | |
| 42 | XZ006 | Orinoco | 烤烟 Flue-cured | 117 | SS113 | 千层塔Qiancengta | 晒烟 Sun-cured | |
| 43 | XZ007 | Mammoth gold | 烤烟 Flue-cured | 118 | SS132 | 腾冲大柳叶 Tengchongdaliuye | 晒烟 Sun-cured | |
| 44 | XZ008 | Golden wilf | 烤烟 Flue-cured | 119 | SS142 | 小叶尖Xiaoyejian | 晒烟 Sun-cured | |
| 45 | XZ009 | CV91 | 烤烟 Flue-cured | 120 | SS146 | 一朵花Yiduohua | 晒烟 Sun-cured | |
| 46 | XZ010 | 云烟315Yunyan 315 | 烤烟 Flue-cured | 121 | SS164 | 大枇杷烟Dapipayan | 晒烟 Sun-cured | |
| 47 | FS027 | 大黄金5210 Dahuangjin 5210 | 烤烟 Flue-cured | 122 | SS169 | 金山乡旱烟 Jinshanxianghanyan | 晒烟 Sun-cured | |
| 48 | FS029 | 单育2号Danyu No.2 | 烤烟 Flue-cured | 123 | XZ031 | K17 | 晒烟 Sun-cured | |
| 49 | FS037 | 广黄55 Guanghuang 55 | 烤烟 Flue-cured | 124 | XZ032 | 寸三皮Cunsanpi | 晒烟 Sun-cured | |
| 50 | FS044 | 红星一号Hongxingyihao | 烤烟 Flue-cured | 125 | XZ033 | 九月寒Jiuyuehan | 晒烟 Sun-cured | |
| 51 | FS045 | 金星6007 Jinxing 6007 | 烤烟 Flue-cured | 126 | XZ034 | 三生烟Sanshengyan | 晒烟 Sun-cured | |
| 52 | FS050 | 净叶黄Jingyehuang | 烤烟 Flue-cured | 127 | BG012 | W.B68 | 白肋烟 Burley | |
| 53 | FS061 | 潘园黄Panyuanhuang | 烤烟 Flue-cured | 128 | BG018 | ky41A | 白肋烟Burley | |
| 54 | FS077 | 许金2号Xujin No. 2 | 烤烟 Flue-cured | 129 | BG023 | 半铁泡Bantiepao | 白肋烟Burley | |
| 55 | FS084 | 云烟87 Yunyan 87 | 烤烟 Flue-cured | 130 | BG029 | BY21 | 白肋烟Burley | |
| 56 | FS085 | 云烟2号Yunyan No.2 | 烤烟 Flue-cured | 131 | BG032 | KY10 | 白肋烟Burley | |
| 57 | FS089 | 中烟90 Zhongyan 90 | 烤烟 Flue-cured | 132 | BG033 | KY14 | 白肋烟Burley | |
| 58 | FY017 | 春雷三号 Chunlei No.3 | 烤烟 Flue-cured | 133 | AG002 | 从江塘洞烟 Congjiangtangdongyan | 晾烟Air-cured | |
| 59 | FY030 | 湄潭黑团壳 Meitanheituanke | 烤烟 Flue-cured | 134 | AG018 | 台江小柳叶 Taijiangxiaoliuye | 晾烟Air-cured | |
| 60 | FS083 | 云烟85 Yunyan 85 | 烤烟 Flue-cured | 135 | AG006 | 剑河乡洞土烟 Jianhexiangdongtuyan | 晾烟Air-cured | |
| 61 | XZ011 | 云烟97 Yunyan 97 | 烤烟 Flue-cured | 136 | GG001 | Ambale-ma | 雪茄烟 Cigar | |
| 62 | FY048 | 兴烟一号Xingyanyihao | 烤烟 Flue-cured | 137 | GG002 | Beinhart1000-1 | 雪茄烟 Cigar | |
| 63 | FT011 | 南江3号Nanjiang No.3 | 烤烟 Flue-cured | 138 | GG004 | Florida301 | 雪茄烟Cigar | |
| 64 | FY068 | 遵烟6号Zunyan No.6 | 烤烟 Flue-cured | 139 | GG005 | Pennbel 69 | 雪茄烟Cigar | |
| 65 | XZ012 | 中烟100 Zhongyan 100 | 烤烟 Flue-cured | 140 | XZ035 | 包皮Baopi | 雪茄烟Cigar | |
| 66 | FS032 | 贵烟3号Guiyan No.3 | 烤烟 Flue-cured | 141 | XZ036 | 坦桑尼亚烟 Tansangniyayan | 香料烟 Oriental | |
| 67 | FS087 | 中烟14 Zhongyan 14 | 烤烟 Flue-cured | 142 | TG002 | Adcook | 香料烟Oriental | |
| 68 | FS032 | 革新3号Gexin No.3 | 烤烟 Flue-cured | 143 | TG004 | Basma536 | 香料烟Oriental | |
| 69 | PG142 | K326LF | 烤烟 Flue-cured | 144 | TG006 | Cekpka | 香料烟Oriental | |
| 70 | FY094 | Q6 | 烤烟 Flue-cured | 145 | TG011 | xanthi-nc | 香料烟Oriental | |
| 71 | XZ013 | 瓮安2号Wengan No.2 | 烤烟 Flue-cured | 146 | XZ037 | YY1 | 药烟 Medicinal | |
| 72 | XZ014 | 2010A4 | 烤烟 Flue-cured | 147 | XZ038 | YY2 | 药烟Medicinal | |
| 73 | XZ015 | 兴烟2号Xingyan No.2 | 烤烟 Flue-cured | 148 | XZ039 | YY4 | 药烟Medicinal | |
| 74 | XZ016 | 99215-1 | 烤烟 Flue-cured | 149 | XZ040 | YY11 | 药烟Medicinal | |
| 75 | XZ017 | PX2 | 烤烟 Flue-cured | 150 | XZ041 | YY15 | 药烟Medicinal |

图2 开发的烟草SNP标记 A:KASP分型检测图,红色点是具有HEX/HEX型等位基因的种质材料,蓝色簇是具有FAM/FAM型等位基因的种质材料,绿色点表示具有HEX/FAM杂合基因型的种质材料, 灰色簇(NTC)是不含模版的空白对照;B:各染色体开发SNP标记数量统计;C:732个SNP位点在染色体上物理位置,右侧为SNP标记的编号,橘色表示该标记为确定的核心标记
Fig. 2 Developed tobacco SNP markers A: Genotyping map of KASP test, red dot is a germplasm with HEX/HEX allele, blue dot is a germplasm with FAM/FAM allele, green dot is a germplasm with HEX/FAM allele, and NTC: blank control without template. B: The number of the SNP marker on each chromosome. C: The physical location of 732 SNPs on chromosome, the numbers on the right are SNP markers ID, and the orange color shows that the SNP is core marker
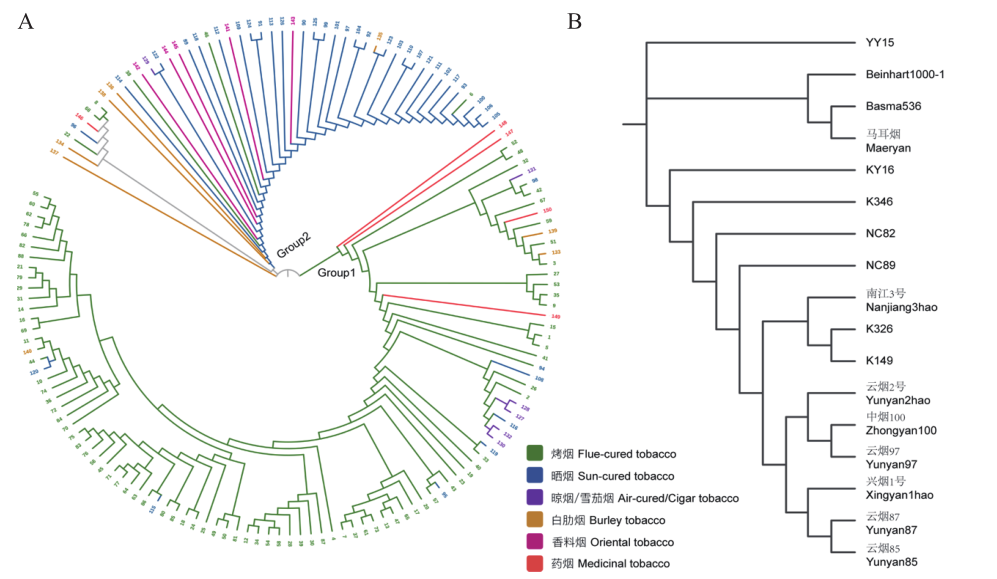
图3 系统发育分析 A:150份供试材料基于48个核心SNP标记系统发育树分析;B:17份代表性烟草种质材料基于48个核心SNP标记系统发育树分析
Fig. 3 Phylogenetic analysis A: The phylogenetic tree of 150 tobacco germplasm based on 48 core SNP markers. B: The phylogenetic tree of 17 representative tobacco cultivars based on 48 core SNP markers
| [1] | 王志德, 张兴伟, 刘艳华. 中国烟草核心种质图谱[M]. 北京: 科学技术文献出版社, 2014. |
| Wang ZD, Zhang XW, Liu YH. Atlas of Chinese tobacco core collection[M]. Beijing: Scientific and Technical Documents Publishing House, 2014. | |
| [2] | 张兴伟. 烟草基因组计划进展篇: 4.中国烟草种质资源平台建设[J]. 中国烟草科学, 2013, 34(4): 112-113. |
| Zhang XW. Tobacco genome project progress: 4. Construction of the China tobacco germplasm platform[J]. Chin Tob Sci, 2013, 34(4): 112-113. | |
| [3] | Frankel O. Genetic perspectives of germplasm conservation[M]// Arber W, et al. Genetics manipulation: Impact on man and society. Cambridge: Cambridge University Press, 1984: 161-170. |
| [4] |
Brown AHD. Core collections: a practical approach to genetic resource management[J]. Genome, 1989, 31(2): 818-824.
doi: 10.1139/g89-144 URL |
| [5] | Brown A, Spillane C. Implementing core collections-principles, procedures, progress, problems and promise[M]// Johnson R, Hodgkin T. Core collections for today and tomorrow. Rome: International Plant Genetic Resources Institute Press, 1999: 1-9. |
| [6] | 董玉琛, 曹永生, 张学勇, 等. 中国普通小麦初选核心种质的产生[J]. 植物遗传资源学报, 2003, 4(1): 1-8. |
| Dong YC, Cao YS, Zhang XY, et al. Establishment of candidate core collections in Chinese common wheat germplasm[J]. J Plant Genet Resour, 2003, 4(1): 1-8. | |
| [7] | 李自超, 张洪亮, 孙传清, 等. 植物遗传资源核心种质研究现状与展望[J]. 中国农业大学学报, 1999, 4(5): 51-62. |
| Li ZC, Zhang HL, Sun CQ, et al. Status and prospects of core collection in plant germplasm resource[J]. J China Agric Univ, 1999, 4(5): 51-62. | |
| [8] | 郭庆法, 高新学. 玉米育种核心种质的构建与有效利用[J]. 中国农业科学, 2000, 33(增刊): 49-56. |
| Guo QF, Gao XX. Construction and effective use of maize core germplasms for breeding[J]. Scientia Agricultura Sinica, 2000, 33(C00): 49-56. | |
| [9] | 周少川, 李宏, 等. 水稻核心种质的育种成效[J]. 中国水稻科学, 2008, 22(1): 51-56. |
| Zhou SC, Li H, et al. Practice and achievement of rice core germplasm breeding[J]. Chin J Rice Sci, 2008, 22(1): 51-56. | |
| [10] | 徐军. 烟草核心种质SSR指纹图谱构建及遗传多样性分析[D]. 北京: 中国农业科学院, 2011. |
| Xu J. Construction of fingerprinting and analysis of genetic diversity with SSR marker for tobacco core germplasm[D]. Beijing: Chinese Academy of Agricultural Sciences, 2011. | |
| [11] | 任民, 等. 基于公共数据库的烟草编码序列SNP位点发掘[C]// 中国作物学会50周年庆祝会暨2011年学术年会论文集. 成都: 中国作物学会, 2011. |
| Ren M, et al. SNP mining for tobacco coding sequences based on public databases[C]// Proceedings of the 50th Anniversary Celebration of the Chinese Crop Society and the 2011 Annual Conference. Chendu: Chinese society of crop sciences, 2011. | |
| [12] | 肖炳光, 邱杰, 曹培健, 等. 利用基因组简约法开发烟草SNP标记及遗传作图[J]. 作物学报, 2014, 40(3): 397-404. |
| Xiao BG, Qiu J, Cao PJ, et al. Development and genetic mapping of SNP markers via genome complexity reduction in tobacco[J]. Acta Agron Sin, 2014, 40(3): 397-404. | |
| [13] | 蔡露, 杨欢, 王勇, 等. 利用GBS技术开发烟草SNP标记及遗传多样性分析[J]. 中国烟草科学, 2018, 39(5): 17-24. |
| Cai L, Yang H, Wang Y, et al. Analysis of genetic diversity of tobacco germplasm resources based on SNP markers via genotyping-by-sequencing technology[J]. Chin Tob Sci, 2018, 39(5): 17-24. | |
| [14] | 任民, 程立锐, 刘旦, 等. 基于RAD重测序技术开发烟草品种SNP位点[J]. 中国烟草科学, 2018, 39(3): 10-17. |
| Ren M, Cheng LR, Liu D, et al. Development of tobacco SNPs based on RAD re-sequencing technology[J]. Chin Tob Sci, 2018, 39(3): 10-17. | |
| [15] | 吴宇瑶. 基于烟草转录组的SNP标记开发及其初步应用[D]. 贵阳: 贵州大学, 2020. |
| Wu YY. Development and preliminary application of SNP markers based on tobacco transcriptome[D]. Guiyang: Guizhou University, 2020. | |
| [16] |
Semagn K, Babu R, Hearne S, et al. Single nucleotide polymorphism genotyping using Kompetitive Allele Specific PCR(KASP): overview of the technology and its application in crop improvement[J]. Mol Breed, 2014, 33(1): 1-14.
doi: 10.1007/s11032-013-9917-x URL |
| [17] | 余世洲, 曹培健, 李泽锋, 等. 基于烟草基因组重测序数据的SNP提取软件组合比较[J]. 烟草科技, 2017, 50(10): 1-7. |
| Yu SZ, Cao PJ, Li ZF, et al. Assessment of SNP-calling pipelines using tobacco genome resequencing data[J]. Tob Sci & Technol, 2017, 50(10): 1-7. | |
| [18] |
Serrote CML, et al. Determining the polymorphism information content of a molecular marker[J]. Gene, 2020, 726: 144175.
doi: 10.1016/j.gene.2019.144175 URL |
| [19] |
Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses[J]. Am J Hum Genet, 2007, 81(3): 559-575.
doi: 10.1086/519795 pmid: 17701901 |
| [20] |
Danecek P, Auton A, Abecasis G, et al. The variant call format and VCF tools[J]. Bioinformatics, 2011, 27(15): 2156-2158.
doi: 10.1093/bioinformatics/btr330 pmid: 21653522 |
| [21] |
Shete S, Tiwari H, Elston RC. On estimating the heterozygosity and polymorphism information content value[J]. Theor Popul Biol, 2000, 57(3): 265-271.
pmid: 10828218 |
| [22] |
Hu H, Liu X, et al. Evaluating information content of SNPs for sample-tagging in re-sequencing projects[J]. Sci Rep, 2015, 5: 10247.
doi: 10.1038/srep10247 pmid: 25975447 |
| [23] |
Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets[J]. Mol Biol Evol, 2016, 33(7): 1870-1874.
doi: 10.1093/molbev/msw054 pmid: 27004904 |
| [24] |
王琰琰, 王俊, 刘国祥, 等. 基于SSR标记的雪茄烟种质资源指纹图谱库的构建及遗传多样性分析[J]. 作物学报, 2021, 47(7): 1259-1274.
doi: 10.3724/SP.J.1006.2021.04183 |
| Wang YY, Wang J, Liu GX, et al. Construction of SSR fingerprint database and genetic diversity analysis of cigar germplasm resources[J]. Acta Agron Sin, 2021, 47(7): 1259-1274. | |
| [25] |
陆海燕, 周玲, 林峰, 等. 基于高通量测序开发玉米高效KASP分子标记[J]. 作物学报, 2019, 45(6): 872-878.
doi: 10.3724/SP.J.1006.2019.83067 |
| Lu HY, Zhou L, Lin F, et al. Development of efficient KASP molecular markers based on high throughput sequencing in maize[J]. Acta Agron Sin, 2019, 45(6): 872-878. | |
| [26] |
李志远, 等. 甘蓝SNP标记开发及主要品种的DNA指纹图谱构建[J]. 中国农业科学, 2018, 51(14): 2771-2788.
doi: 10.3864/j.issn.0578-1752.2018.14.014 |
| Li ZY, et al. Development of SNP markers in cabbage and construction of DNA fingerprinting of main varieties[J]. Sci Agric Sin, 2018, 51(14): 2771-2788. | |
| [27] | 郑国彬, 赵虹博, 黄良敏, 等. 黄鳍棘鲷SNP标记开发与验证[J/OL]. 热带海洋学报, 2022. http://www.jto.ac.cn/CN/10.11978/2022108. |
| Zhen GB, Zhao HB, Huang LM, et al. Discovery and verification of SNP in Acabthopagrus latus[J/OL]. J Trop Oceanogr, 2022. http://www.jto.ac.cn/CN/10.11978/2022108. | |
| [28] |
杨青青, 唐家琪, 张昌泉, 等. KASP标记技术在主要农作物中的应用及展望[J]. 生物技术通报, 2022, 38(4): 58-71.
doi: 10.13560/j.cnki.biotech.bull.1985.2021-1378 |
| Yang QQ, Tang JQ, Zhang CQ, et al. Application and prospect of KASP marker technology in main crops[J]. Biotechnol Bull, 2022, 38(4): 58-71. | |
| [29] | 尹国英, 杨小燕, 等. 鉴定烟草种质资源SSR核心引物筛选和验证[J]. 植物遗传资源学报, 2013, 14(5): 960-965. |
| Yin GY, Yang XY, et al. Screen and identification of SSR core primers for tobacco germplasm[J]. J Plant Genet Resour, 2013, 14(5): 960-965. | |
| [30] | 张铭真, 邢雪霞, 李晓辉, 等. 烟草种质资源遗传多样性与群体结构分析[J]. 中国烟草科学, 2016, 37(4): 42-47. |
| Zhang MZ, Xing XX, Li XH, et al. Analysis of genetic diversity and population structure of germplasm resources in Nicotiana tabacum[J]. Chin Tob Sci, 2016, 37(4): 42-47. | |
| [31] | 王琰琰. 雪茄烟种质资源SNP指纹图谱构建及群体遗传分析[D]. 北京: 中国农业科学院, 2021. |
| Wang YY. Construction of a SNP fingerprinting database and population genetic analysis of cigar tobacco germplasm resources in China[D]. Beijing: Chinese Academy of Agricultural Sciences, 2021. |
| [1] | 杨志晓, 侯骞, 刘国权, 卢志刚, 曹毅, 芶剑渝, 王轶, 林英超. 不同抗性烟草品系Rubisco及其活化酶对赤星病胁迫的响应[J]. 生物技术通报, 2023, 39(9): 202-212. |
| [2] | 刘珍银, 段郅臻, 彭婷, 王童欣, 王健. 基于三角梅的病毒诱导基因沉默体系的建立与优化[J]. 生物技术通报, 2023, 39(7): 123-130. |
| [3] | 李文辰, 刘鑫, 康越, 李伟, 齐泽铮, 于璐, 王芳. TRV病毒诱导大豆基因沉默体系优化及应用[J]. 生物技术通报, 2023, 39(7): 143-150. |
| [4] | 张路阳, 韩文龙, 徐晓雯, 姚健, 李芳芳, 田效园, 张智强. 烟草TCP基因家族的鉴定及表达分析[J]. 生物技术通报, 2023, 39(6): 248-258. |
| [5] | 王羽, 尹铭绅, 尹晓燕, 奚家勤, 杨建伟, 牛秋红. 烟草甲体内烟碱降解菌的筛选、鉴定及降解特性[J]. 生物技术通报, 2023, 39(6): 308-315. |
| [6] | 申云鑫, 施竹凤, 周旭东, 李铭刚, 张庆, 冯路遥, 陈齐斌, 杨佩文. 三株具生防功能芽孢杆菌的分离鉴定及其生物活性研究[J]. 生物技术通报, 2023, 39(3): 267-277. |
| [7] | 蔡梦鲜, 高作敏, 胡利娟, 冯群, 王洪程, 朱斌. 天然甘蓝型油菜C染色体组C1,C2缺体的创建及遗传分析[J]. 生物技术通报, 2023, 39(3): 81-88. |
| [8] | 杜清洁, 周璐瑶, 杨思震, 张嘉欣, 陈春林, 李娟起, 李猛, 赵士文, 肖怀娟, 王吉庆. 过表达CaCP1提高转基因烟草对盐胁迫的敏感性[J]. 生物技术通报, 2023, 39(2): 172-182. |
| [9] | 汪格格, 邱诗蕊, 张琳晗, 杨国伟, 徐小云, 汪爱羚, 曾淑华, 刘雅洁. 异源三倍体普通烟草(SST)减数分裂期的分子细胞学研究[J]. 生物技术通报, 2023, 39(2): 183-192. |
| [10] | 车永梅, 刘广超, 郭艳苹, 叶青, 赵方贵, 刘新. 一种耐盐复合菌剂的制备和促生作用研究[J]. 生物技术通报, 2023, 39(11): 217-225. |
| [11] | 尹国英, 刘畅, 常永春, 羽王洁, 王兵, 张盼, 郭玉双. 烟草半胱氨酸蛋白酶家族和相应miRNAs的鉴定及其对PVY的响应[J]. 生物技术通报, 2023, 39(10): 184-196. |
| [12] | 刘广超, 叶青, 车永梅, 李雅华, 安东, 刘新. 烟草根际高效解磷菌的筛选鉴定及促生作用研究[J]. 生物技术通报, 2022, 38(8): 179-187. |
| [13] | 张昊鑫, 王中华, 牛兵, 郭慷, 刘璐, 姜瑛, 张仕祥. 产IAA兼具溶磷解钾高效促生菌的筛选、鉴定及其广谱性应用[J]. 生物技术通报, 2022, 38(5): 100-111. |
| [14] | 付偲僮, 司未佳, 刘颖, 程堂仁, 王佳, 张启翔, 潘会堂. TRV介导的小报春基因沉默技术体系的建立[J]. 生物技术通报, 2022, 38(4): 295-302. |
| [15] | 邹雪峰, 李铭刚, 包玲风, 陈齐斌, 赵江源, 汪林, 濮永瑜, 郝大程, 张庆, 杨佩文. 一株分泌型铁载体真菌分离鉴定及生物活性研究[J]. 生物技术通报, 2022, 38(3): 130-138. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||