








生物技术通报 ›› 2023, Vol. 39 ›› Issue (3): 101-115.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0851
宋海娜1( ), 吴心桐1, 杨鲁豫1, 耿喜宁1, 张华敏2(
), 吴心桐1, 杨鲁豫1, 耿喜宁1, 张华敏2( ), 宋小龙3
), 宋小龙3
收稿日期:2022-07-10
出版日期:2023-03-26
发布日期:2023-04-10
通讯作者:
张华敏,男,博士,讲师,研究方向:植物遗传与分子育种;E-mail: hmzhang111@126.com作者简介:宋海娜,女,博士,讲师,研究方向:植物分子育种;E-mail: shn1126@126.com
基金资助:
SONG Hai-na1( ), WU Xin-tong1, YANG Lu-yu1, GENG Xi-ning1, ZHANG Hua-min2(
), WU Xin-tong1, YANG Lu-yu1, GENG Xi-ning1, ZHANG Hua-min2( ), SONG Xiao-long3
), SONG Xiao-long3
Received:2022-07-10
Published:2023-03-26
Online:2023-04-10
摘要:
葱鳞葡萄胞菌引起的韭菜灰霉病是影响韭菜产量和品质的主要因素之一。为了筛选出感染灰霉病后韭菜叶片中稳定表达的内参基因用于基因定量表达分析,以模拟接种和接种葱鳞葡萄孢菌24、48、72 h的韭菜叶片为材料,基于前期的转录组测序结果选取 UBC1、UBC2、UBQ1、UBQ2、GAPDH3、GAPDH4、TUB、EF-1α、40S RP、DDX、eIF-1A、PABP和DnaJ共13个基因为候选内参基因,利用实时荧光定量PCR(RT-qPCR)技术检测13个基因的表达情况,采用geNorm、NormFinder、BestKeeper软件和Reffinder在线程序对候选内参基因的表达稳定性进行评估。结果表明,13个候选内参基因中UBQ1的Ct值变化范围最小,表达水平最稳定。GeNorm、NormFinder和BestKeeper 软件筛选出的最佳内参基因不同,RefFinder综合评估显示,UBC2和UBQ1是韭菜叶片接种葱鳞葡萄孢菌后表达稳定性较好的基因,DDX是稳定性较差的基因。为了验证所筛选内参基因的可靠性,选择6个稳定性不同的候选内参基因分别作为定量分析的内部参照,对接种葱鳞葡萄孢菌后不同时间韭菜叶片中GST676和PRP902基因的表达水平进行归一化处理。结果显示,以稳定性较高的UBC2或UBQ1为内参基因时,校正的GST676和PRP902基因的表达趋势与转录组测序结果一致,而以稳定性相对较差的40S RP、eIF-1A、GAPDH4或DDX为内参基因时,校正的目的基因的表达趋势与转录组测序结果稍有差异。软件分析和实验验证结果表明,UBC2和UBQ1基因可作为葱鳞葡萄孢菌侵染韭菜叶片后相关基因定量表达分析的最合适内参基因。研究结果为后续开展韭菜抗灰霉病关键基因表达分析和功能研究奠定了基础。
宋海娜, 吴心桐, 杨鲁豫, 耿喜宁, 张华敏, 宋小龙. 葱鳞葡萄孢菌诱导下韭菜RT-qPCR内参基因的筛选和验证[J]. 生物技术通报, 2023, 39(3): 101-115.
SONG Hai-na, WU Xin-tong, YANG Lu-yu, GENG Xi-ning, ZHANG Hua-min, SONG Xiao-long. Selection and Validation of Reference Genes for RT-qPCR in Allium tuberosum Infected by Botrytis squamosa[J]. Biotechnology Bulletin, 2023, 39(3): 101-115.
| 基因名称Gene symbol | 基因ID Gene ID | 引物序列Primer sequence(5'-3') | 扩增长度Amplified length/bp |
|---|---|---|---|
| UBC1 | F02_cb4795_c6/f1p0/677 | F:CACTTCCCTCCTGACTATCCCT R:CAAATACTGCCATTGCTGTTGA | 92 |
| UBC2 | F02_cb10194_c5/f1p0/683 | F:AAGGTGCTCCTGTCTATCTGCT R:TGATTCATACTTGGCTCTGTCG | 108 |
| UBQ1 | F02_cb4331_c39/f4p2/2064 | F:GTCAACTCCGTCACCGTAAACA R:GCAACTTCGGACTTCTCCTCTA | 119 |
| UBQ2 | F02_cb3159_c105/f1p2/1236 | F:ACCTTGTGCTCCGTCTTCGTG R:AACATCTGGCTTACCGTCGTCT | 162 |
| GAPDH3 | F02_cb2521_c25/f1p0/1405 | F:GACAAGCCAGTTGCTGTATTCG R:CTTTGCACCACCCTTAATGTGA | 140 |
| GAPDH4 | F02_cb2521_c44/f7p1/1415 | F:AATCACTGCCACCCAGAAGACT R:ACGGAATGACATGCCAGTAAGC | 163 |
| TUB | F02_cb8195_c2075/f1p0/1651 | F:GGTGGTGGTACTGGATCTGG R:TTGTATGGCTCCACAACTGC | 134 |
| EF-1α | F02_cb8884_c10/f1p3/1571 | F:ACCGAGTCCATTCCTCCTGTTC R:GGGTTTAGGTGCCTCCTCTTCA | 147 |
| 40S RP | F02_cb13441_c1/f3p0/558 | F:TGACCTCTTGAATCCACCTGC R:AGCAACCTTGGCACTTGACA | 107 |
| PABP | F02_cb2343_c66/f1p0/2490 | F:TCCTGATATGGCTGGCTTGC R:CCTCGGGCGATTCAAGAAGA | 224 |
| DnaJ | F02_cb2629_c9/f1p0/682 | F:AGACTCCCTGCAATTCCAGC R:TCTTCATGAACTCCTCGGCG | 223 |
| DDX | F02_cb3613_c7/f1p2/2466 | F:CCATGTTCAAGAGCAGCACG R:CATGAAGAGCACGAGACCCA | 145 |
| eIF-1A | F02_cb13117_c3/f2p1/651 | F:GGCTTTGTCATATTCGGGGC R:ATCACCAGCACCATCGTCC | 227 |
| GST676 | F02_cb3237_c0/f37p2/676 | F:GCAAGTCGGTAAAGCTCGGA R:ACTGGCCAAAAGAGTAGCGA | 160 |
| PRP902 | F02_cb14527_c723/f1p0/902 | F:CGTCACAGACAAGCAAAGGC R:CTTACTGTGCCACGTGGGAT | 180 |
表1 基因和引物序列信息
Table 1 Genes and primer sequences
| 基因名称Gene symbol | 基因ID Gene ID | 引物序列Primer sequence(5'-3') | 扩增长度Amplified length/bp |
|---|---|---|---|
| UBC1 | F02_cb4795_c6/f1p0/677 | F:CACTTCCCTCCTGACTATCCCT R:CAAATACTGCCATTGCTGTTGA | 92 |
| UBC2 | F02_cb10194_c5/f1p0/683 | F:AAGGTGCTCCTGTCTATCTGCT R:TGATTCATACTTGGCTCTGTCG | 108 |
| UBQ1 | F02_cb4331_c39/f4p2/2064 | F:GTCAACTCCGTCACCGTAAACA R:GCAACTTCGGACTTCTCCTCTA | 119 |
| UBQ2 | F02_cb3159_c105/f1p2/1236 | F:ACCTTGTGCTCCGTCTTCGTG R:AACATCTGGCTTACCGTCGTCT | 162 |
| GAPDH3 | F02_cb2521_c25/f1p0/1405 | F:GACAAGCCAGTTGCTGTATTCG R:CTTTGCACCACCCTTAATGTGA | 140 |
| GAPDH4 | F02_cb2521_c44/f7p1/1415 | F:AATCACTGCCACCCAGAAGACT R:ACGGAATGACATGCCAGTAAGC | 163 |
| TUB | F02_cb8195_c2075/f1p0/1651 | F:GGTGGTGGTACTGGATCTGG R:TTGTATGGCTCCACAACTGC | 134 |
| EF-1α | F02_cb8884_c10/f1p3/1571 | F:ACCGAGTCCATTCCTCCTGTTC R:GGGTTTAGGTGCCTCCTCTTCA | 147 |
| 40S RP | F02_cb13441_c1/f3p0/558 | F:TGACCTCTTGAATCCACCTGC R:AGCAACCTTGGCACTTGACA | 107 |
| PABP | F02_cb2343_c66/f1p0/2490 | F:TCCTGATATGGCTGGCTTGC R:CCTCGGGCGATTCAAGAAGA | 224 |
| DnaJ | F02_cb2629_c9/f1p0/682 | F:AGACTCCCTGCAATTCCAGC R:TCTTCATGAACTCCTCGGCG | 223 |
| DDX | F02_cb3613_c7/f1p2/2466 | F:CCATGTTCAAGAGCAGCACG R:CATGAAGAGCACGAGACCCA | 145 |
| eIF-1A | F02_cb13117_c3/f2p1/651 | F:GGCTTTGTCATATTCGGGGC R:ATCACCAGCACCATCGTCC | 227 |
| GST676 | F02_cb3237_c0/f37p2/676 | F:GCAAGTCGGTAAAGCTCGGA R:ACTGGCCAAAAGAGTAGCGA | 160 |
| PRP902 | F02_cb14527_c723/f1p0/902 | F:CGTCACAGACAAGCAAAGGC R:CTTACTGTGCCACGTGGGAT | 180 |
| 基因 名称 Gene symbol | 模拟接菌不同时间的样品 Mock-inoculated samples | 接种葱鳞葡萄孢菌不同时间的样品 Samples inoculated with B. squamosa | ||||||
|---|---|---|---|---|---|---|---|---|
| 24 h | 48 h | 72 h | 24 h | 48 h | 72 h | |||
| UBC1 | 14.63 | 14.47 | 15.48 | 21.21 | 14.66 | 21.60 | ||
| UBC2 | 45.80 | 53.35 | 59.64 | 72.14 | 51.73 | 83.00 | ||
| UBQ1 | 8.75 | 7.90 | 10.06 | 12.67 | 8.00 | 10.81 | ||
| UBQ2 | 187.15 | 189.18 | 197.98 | 223.79 | 139.12 | 245.41 | ||
| GAPDH3 | 17.86 | 28.28 | 23.98 | 25.81 | 27.96 | 15.07 | ||
| GAPDH4 | 111.50 | 168.16 | 154.12 | 214.92 | 132.19 | 155.86 | ||
| TUB | 49.37 | 58.77 | 50.06 | 58.57 | 52.65 | 47.37 | ||
| EF-1α | 28.34 | 18.20 | 26.84 | 48.63 | 13.83 | 22.83 | ||
| 40S RP | 183.95 | 252.63 | 198.60 | 206.80 | 235.64 | 163.86 | ||
| PABP | 87.77 | 97.44 | 88.98 | 97.07 | 97.91 | 72.27 | ||
| DnaJ | 516.66 | 480.78 | 478.24 | 429.37 | 332.07 | 522.24 | ||
| DDX | 48.00 | 43.42 | 35.19 | 51.43 | 38.59 | 33.68 | ||
| eIF-1A | 31.35 | 44.60 | 34.58 | 38.97 | 33.39 | 33.45 | ||
表2 候选内参基因在RNA-Seq中的FPKM值
Table 2 FPKM values of candidate reference genes in the samples by RNA-Seq
| 基因 名称 Gene symbol | 模拟接菌不同时间的样品 Mock-inoculated samples | 接种葱鳞葡萄孢菌不同时间的样品 Samples inoculated with B. squamosa | ||||||
|---|---|---|---|---|---|---|---|---|
| 24 h | 48 h | 72 h | 24 h | 48 h | 72 h | |||
| UBC1 | 14.63 | 14.47 | 15.48 | 21.21 | 14.66 | 21.60 | ||
| UBC2 | 45.80 | 53.35 | 59.64 | 72.14 | 51.73 | 83.00 | ||
| UBQ1 | 8.75 | 7.90 | 10.06 | 12.67 | 8.00 | 10.81 | ||
| UBQ2 | 187.15 | 189.18 | 197.98 | 223.79 | 139.12 | 245.41 | ||
| GAPDH3 | 17.86 | 28.28 | 23.98 | 25.81 | 27.96 | 15.07 | ||
| GAPDH4 | 111.50 | 168.16 | 154.12 | 214.92 | 132.19 | 155.86 | ||
| TUB | 49.37 | 58.77 | 50.06 | 58.57 | 52.65 | 47.37 | ||
| EF-1α | 28.34 | 18.20 | 26.84 | 48.63 | 13.83 | 22.83 | ||
| 40S RP | 183.95 | 252.63 | 198.60 | 206.80 | 235.64 | 163.86 | ||
| PABP | 87.77 | 97.44 | 88.98 | 97.07 | 97.91 | 72.27 | ||
| DnaJ | 516.66 | 480.78 | 478.24 | 429.37 | 332.07 | 522.24 | ||
| DDX | 48.00 | 43.42 | 35.19 | 51.43 | 38.59 | 33.68 | ||
| eIF-1A | 31.35 | 44.60 | 34.58 | 38.97 | 33.39 | 33.45 | ||
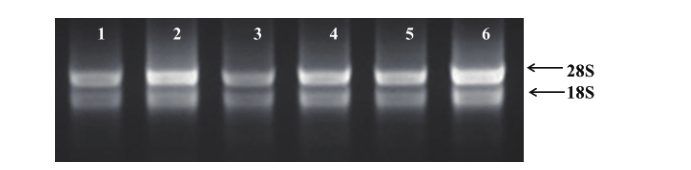
图1 韭菜叶片提取的总RNA 1-3分别是模拟接种24、48和72 h的样品;4-6分别是接种24、48和72 h的样品
Fig. 1 Extracted total RNA in the leaves of Chinese chive A. tvberosum 1-3 are the mock-inoculated samples at 24, 48 and 72 h post inoculation respectively. 4-6 are the samples inoculated with B. squamosa at 24, 48 and 72 h post inoculation respectively

图2 韭菜候选内参基因PCR 扩增产物的琼脂糖凝胶电泳检测
Fig. 2 Agarose electrophoresis analysis of PCR amplified products of candidate reference genes in Chinese chive 1: UBC1; 2: UBC2; 3: UBQ1; 4: UBQ2; 5: GAPDH3; 6: GAPDH4; 7: TUB; 8: EF-1a; 9: 40S RP; 10: DDX; 11: eIF-1A; 12: PABP; 13: DnaJ

图3 韭菜候选内参基因实时荧光定量PCR扩增的熔解曲线 -dF/dT是荧光对温度的一阶负导数
Fig. 3 Melting curves of candidate reference genes in Chinese chive for RT-qPCR amplification -dF/dT is the first negative derivative of fluorescence(F)with respect to temperature(T)
| 基因名称 Gene symbol | 线性相关系数Linear correlation coefficient(R2) | 扩增效率Amplification efficiency/% |
|---|---|---|
| UBC1 | 0.993 9 | 104.83 |
| UBC2 | 0.963 0 | 101.12 |
| UBQ1 | 0.994 1 | 110.70 |
| UBQ2 | 0.990 7 | 106.64 |
| GAPDH3 | 0.995 5 | 109.88 |
| GAPDH4 | 0.996 6 | 110.65 |
| EF-1α | 0.991 2 | 110.36 |
| TUB | 0.991 7 | 107.54 |
| 40S RP | 0.995 3 | 100.80 |
| DDX | 0.990 3 | 99.19 |
| eIF-1A | 0.997 5 | 106.30 |
| PABP | 0.990 9 | 96.06 |
| DnaJ | 0.998 9 | 106.97 |
表3 候选内参基因扩增效率分析
Table 3 Amplification efficiency of candidate reference genes
| 基因名称 Gene symbol | 线性相关系数Linear correlation coefficient(R2) | 扩增效率Amplification efficiency/% |
|---|---|---|
| UBC1 | 0.993 9 | 104.83 |
| UBC2 | 0.963 0 | 101.12 |
| UBQ1 | 0.994 1 | 110.70 |
| UBQ2 | 0.990 7 | 106.64 |
| GAPDH3 | 0.995 5 | 109.88 |
| GAPDH4 | 0.996 6 | 110.65 |
| EF-1α | 0.991 2 | 110.36 |
| TUB | 0.991 7 | 107.54 |
| 40S RP | 0.995 3 | 100.80 |
| DDX | 0.990 3 | 99.19 |
| eIF-1A | 0.997 5 | 106.30 |
| PABP | 0.990 9 | 96.06 |
| DnaJ | 0.998 9 | 106.97 |
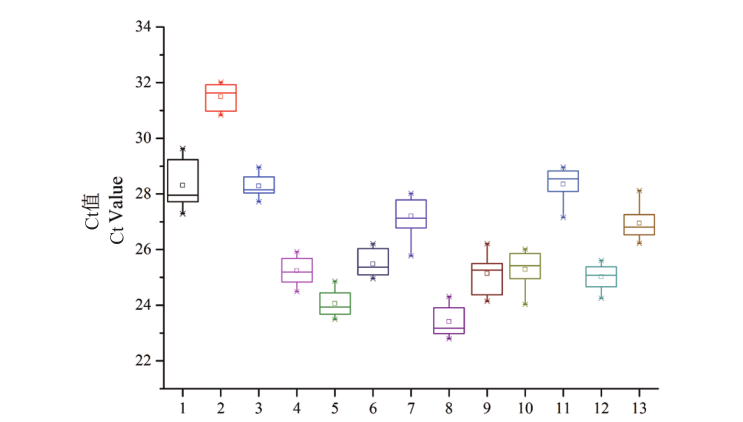
图4 候选内参基因在所有样品中的Ct值分布 方框内的横线表示Ct值的平均值
Fig. 4 Distribution of Ct values of candidate reference genes across all samples The horizontal line in the box indicates the average Ct values. 1: EF-1α; 2: UBQ1; 3: TUB; 4: UBQ2; 5: 40S RP; 6: eIF-1A; 7: DnaJ; 8: PABP; 9: DDX; 10: GAPDH4; 11: UBC1; 12: UBC2; 13: GAPDH3
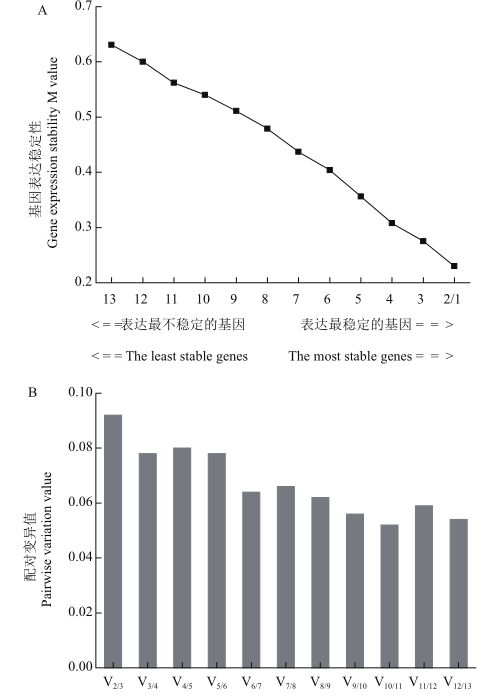
图5 geNorm软件分析候选内参基因的表达稳定性 A:候选内参基因表达稳定性分析;B:归一化内参基因数目的确定
Fig. 5 Expression stabilities of candidate reference genes assayed by geNorm software A is the average expression stability M value of candidate reference genes. The least stable genes are on the left and the most stable genes on the right. B is the normalized number of reference genes. In figure A, 1: eIF-1A; 2: UBQ2; 3: UBC2; 4: 40S RP; 5: PABP; 6: UBC1; 7: UBQ1; 8: DnaJ; 9: GAPDH4; 10: TUB; 11: GAPDH3; 12: EF-1a; 13: DDX
| 基因名称Gene symbol | 稳定值 Stability | 排名Rank |
|---|---|---|
| UBQ1 | 0.195 | 1 |
| UBC2 | 0.261 | 2 |
| 40S RP | 0.28 | 3 |
| UBC1 | 0.374 | 4 |
| UBQ2 | 0.375 | 5 |
| eIF-1A | 0.443 | 6 |
| GAPDH4 | 0.456 | 7 |
| TUB | 0.478 | 8 |
| PABP | 0.48 | 9 |
| GAPDH3 | 0.51 | 10 |
| DnaJ | 0.581 | 11 |
| EF-1α | 0.642 | 12 |
| DDX | 0.654 | 13 |
表4 NormFinder软件分析候选内参基因的表达稳定性
Table 4 Expression stabilities of candidate reference genes assayed by NormFinder software
| 基因名称Gene symbol | 稳定值 Stability | 排名Rank |
|---|---|---|
| UBQ1 | 0.195 | 1 |
| UBC2 | 0.261 | 2 |
| 40S RP | 0.28 | 3 |
| UBC1 | 0.374 | 4 |
| UBQ2 | 0.375 | 5 |
| eIF-1A | 0.443 | 6 |
| GAPDH4 | 0.456 | 7 |
| TUB | 0.478 | 8 |
| PABP | 0.48 | 9 |
| GAPDH3 | 0.51 | 10 |
| DnaJ | 0.581 | 11 |
| EF-1α | 0.642 | 12 |
| DDX | 0.654 | 13 |
| 基因名称Gene symbol | 标准偏差Standard deviation(SD) | 变异系数Coefficient of variation(CV) | 调节系数标准差Std dev[± x-fold] | 排名Rank |
|---|---|---|---|---|
| TUB | 0.327 | 1.156 | 1.254 | 1 |
| UBC2 | 0.390 | 1.560 | 1.311 | 2 |
| 40S RP | 0.396 | 1.647 | 1.316 | 3 |
| eIF-1A | 0.411 | 1.615 | 1.330 | 4 |
| UBQ1 | 0.416 | 1.320 | 1.334 | 5 |
| PABP | 0.435 | 1.856 | 1.352 | 6 |
| UBQ2 | 0.436 | 1.726 | 1.353 | 7 |
| UBC1 | 0.472 | 1.666 | 1.387 | 8 |
| GAPDH3 | 0.477 | 1.768 | 1.391 | 9 |
| DnaJ | 0.494 | 1.811 | 1.408 | 10 |
| DDX | 0.560 | 2.225 | 1.474 | 11 |
| GAPDH4 | 0.596 | 2.359 | 1.512 | 12 |
| EF-1α | 0.727 | 2.567 | 1.655 | 13 |
表5 BestKeeper软件分析候选内参基因的表达稳定性
Table 5 Expression stabilities of candidate reference genes assayed by BestKeeper software
| 基因名称Gene symbol | 标准偏差Standard deviation(SD) | 变异系数Coefficient of variation(CV) | 调节系数标准差Std dev[± x-fold] | 排名Rank |
|---|---|---|---|---|
| TUB | 0.327 | 1.156 | 1.254 | 1 |
| UBC2 | 0.390 | 1.560 | 1.311 | 2 |
| 40S RP | 0.396 | 1.647 | 1.316 | 3 |
| eIF-1A | 0.411 | 1.615 | 1.330 | 4 |
| UBQ1 | 0.416 | 1.320 | 1.334 | 5 |
| PABP | 0.435 | 1.856 | 1.352 | 6 |
| UBQ2 | 0.436 | 1.726 | 1.353 | 7 |
| UBC1 | 0.472 | 1.666 | 1.387 | 8 |
| GAPDH3 | 0.477 | 1.768 | 1.391 | 9 |
| DnaJ | 0.494 | 1.811 | 1.408 | 10 |
| DDX | 0.560 | 2.225 | 1.474 | 11 |
| GAPDH4 | 0.596 | 2.359 | 1.512 | 12 |
| EF-1α | 0.727 | 2.567 | 1.655 | 13 |

图 6 RefFinder分析候选内参基因的表达稳定性
Fig. 6 Expression stabilities of candidate reference genes assayed by RefFinder 1: UBC2; 2: UBQ1; 3: 40S RP; 4: UBQ2; 5: eIF-1A; 6: TUB; 7: UBC1; 8: PABP;9: GAPDH4; 10: GAPDH3; 11: DnaJ; 12: EF-1α; 13: DDX
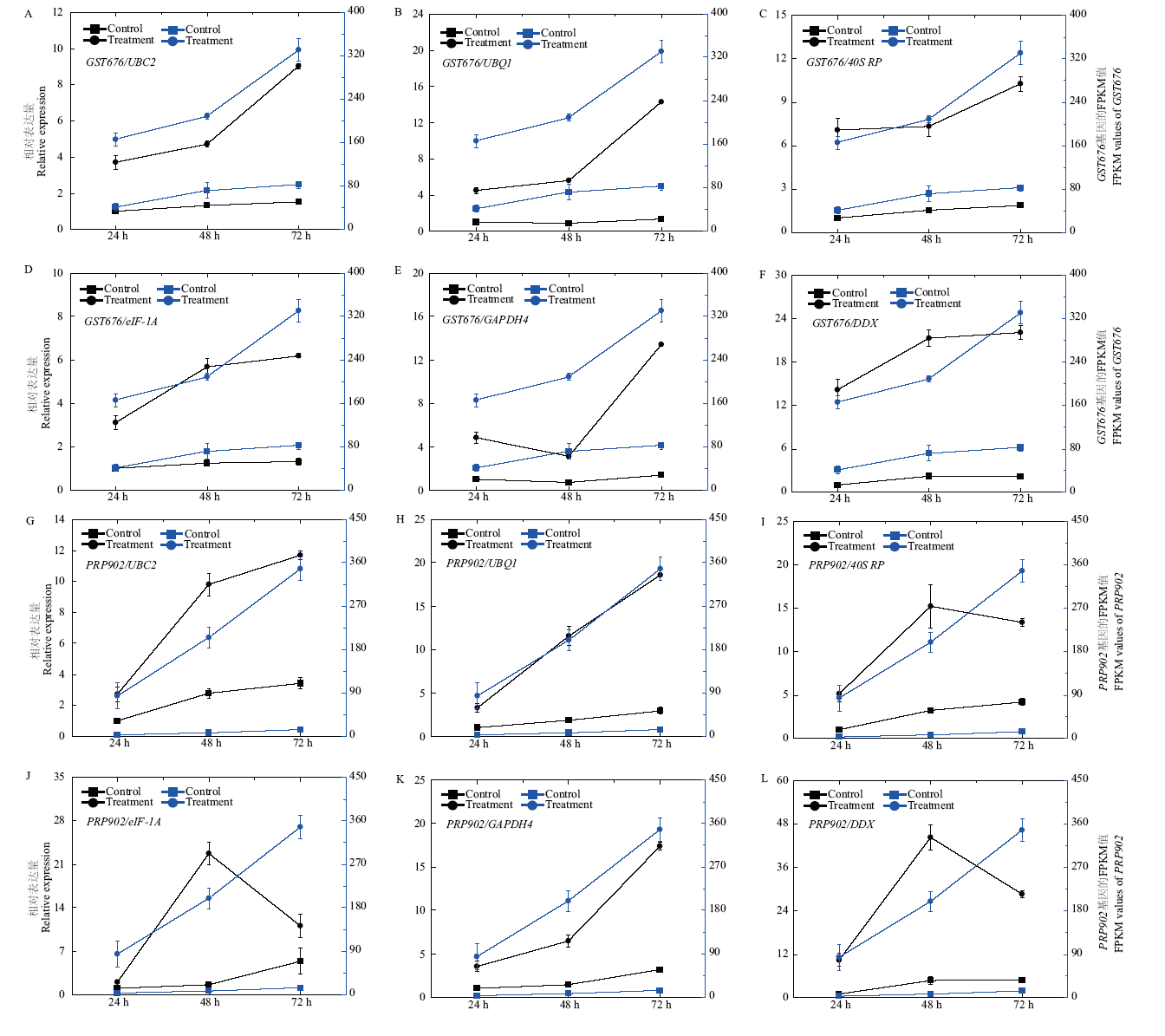
图7 GST676和PRP902基因分别以UBC2、UBQ1、40S RP、eIF-1A、GAPDH4和DDX为内参基因时在模拟接种和接种不同时间韭菜叶片中的相对表达量及其在转录组测序中的FPKM值 Y轴左侧是基因的相对表达量,右侧是基因的FPKM值
Fig. 7 Relative expressions of GST676 and PRP902 in Chinese chive leaves suffered by mock-inoculated and inoculated with B. squamosa when UBC2, UBQ1, 40S RP, eIF-1A, GAPDH4 or DDX used as the internal reference gene, and the FPKM values of GST676 and PRP902 in the samples by RNA-Seq The left side of Y axis is the relative expression, the right side of Y axis is the FPKM values of genes
| [1] | 黄征. 葱蒜类蔬菜真菌病害调查及其病原鉴定[J]. 中国农学通报, 2007, 23(5): 326-329. |
|
Huang Z. Diagnosis and identification on the fungal disease of bulb crops in Fujian[J]. Chin Agric Sci Bull, 2007, 23(5): 326-329.
doi: 10.11924/j.issn.1000-6850.0705326 |
|
| [2] | 崔蕴刚, 张华敏, 李延龙, 等. 韭菜灰霉病病原鉴定及其生物学特性[J]. 北方园艺, 2020(4): 14-19. |
| Cui YG, Zhang HM, Li YL, et al. Identification and biological characterization of the pathogen of Allium tuberosum grey mold[J]. North Hortic, 2020(4): 14-19. | |
| [3] |
Zhang J, Zhang L, Li GQ, et al. Botrytis sinoallii: a new species of the grey mould pathogen on Allium crops in China[J]. Mycoscience, 2010, 51(6): 421-431.
doi: 10.47371/mycosci.MYC51421 URL |
| [4] | 胡彬, 黄中乔, 刘西莉, 等. 9种杀菌剂对韭菜灰霉病的防治效果[J]. 中国农学通报, 2014, 30(4): 293-298. |
|
Hu B, Huang ZQ, Liu XL, et al. Control efficacy of 9 fungicides against Chinese chives gray mold[J]. Chin Agric Sci Bull, 2014, 30(4): 293-298.
doi: 10.11924/j.issn.1000-6850.2013-3021 |
|
| [5] | 胡彬, 李琳, 戚如诗, 等. 从韭菜腐霉利残留超标看农药登记及最大残留限量标准的科学制定[J]. 中国蔬菜, 2020(5): 9-11. |
| Hu B, Li L, Qi RS, et al. The scientific establishment of pesticide registration and maximum residue limit standard from the point of Chinese chives procymidone residue exceed the standard[J]. China Veg, 2020(5): 9-11. | |
| [6] | 高皓杰, 张兰云, 李桐桐, 等. 韭菜灰霉病防治烟剂的筛选与评价[J]. 农药学学报, 2022, 24(2): 315-325. |
| Gao HJ, Zhang LY, Li TT, et al. Screening and evaluation of smoke generators to control gray mold of Chinese chives[J]. Chin J Pestic Sci, 2022, 24(2): 315-325. | |
| [7] | 韩之琪, 贲海燕, 谢学文, 等. 灰葡萄孢对杀菌剂抗性研究进展[J]. 中国蔬菜, 2014(5): 6-10. |
| Han ZQ, Ben HY, Xie XW, et al. Research progress on Botrytis cinerea resistance to different fungicides[J]. China Veg, 2014(5): 6-10. | |
| [8] | Rupp S, Plesken C, Rumsey S, et al. Botrytis fragariae, a new species causing gray mold on strawberries, shows high frequencies of specific and efflux-based fungicide resistance[J]. Appl Environ Microbiol, 2017, 83(9): e00269-e00217. |
| [9] | 胡适宜. 被子植物胚胎学[M]. 北京: 人民教育出版社, 1982. |
| Hu SY. Angiosperm embryology[M]. Beijing: People’s Education Press, 1982. | |
| [10] | 张华敏, 陈建华, 尹守恒, 等. 韭菜无融合生殖种子形成机制研究进展[J]. 河南农业科学, 2020, 49(6): 1-7. |
| Zhang HM, Chen JH, Yin SH, et al. Research progress of the formation mechanism of apomictic seed in Chinese chive[J]. J Henan Agric Sci, 2020, 49(6): 1-7. | |
| [11] |
Gachon C, Mingam A, Charrier B. Real-time PCR: what relevance to plant studies?[J]. J Exp Bot, 2004, 55(402): 1445-1454.
doi: 10.1093/jxb/erh181 pmid: 15208338 |
| [12] |
Nolan T, Hands RE, Bustin SA. Quantification of mRNA using real-time RT-PCR[J]. Nat Protoc, 2006, 1(3): 1559-1582.
doi: 10.1038/nprot.2006.236 pmid: 17406449 |
| [13] |
Dheda K, Huggett JF, Chang JS, et al. The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization[J]. Anal Biochem, 2005, 344(1): 141-143.
doi: 10.1016/j.ab.2005.05.022 pmid: 16054107 |
| [14] |
袁伟, 万红建, 杨悦俭. 植物实时荧光定量PCR内参基因的特点及选择[J]. 植物学报, 2012, 47(4): 427-436.
doi: 10.3724/SP.J.1259.2012.00427 |
| Yuan W, Wan HJ, Yang YJ. Characterization and selection of reference genes for real-time quantitative RT-PCR of plants[J]. Chin Bull Bot, 2012, 47(4): 427-436. | |
| [15] |
Brunner AM, Yakovlev IA, Strauss SH. Validating internal controls for quantitative plant gene expression studies[J]. BMC Plant Biol, 2004, 4: 14.
pmid: 15317655 |
| [16] |
Huggett J, Dheda K, Bustin S, et al. Real-time RT-PCR normalisation; strategies and considerations[J]. Genes Immun, 2005, 6(4): 279-284.
doi: 10.1038/sj.gene.6364190 pmid: 15815687 |
| [17] |
Jain M, Nijhawan A, Tyagi AK, et al. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR[J]. Biochem Biophys Res Commun, 2006, 345(2): 646-651.
doi: 10.1016/j.bbrc.2006.04.140 URL |
| [18] |
蒋婷婷, 高燕会, 童再康. 石蒜属植物实时荧光定量PCR内参基因的选择[J]. 园艺学报, 2015, 42(6): 1129-1138.
doi: 10.16420/j.issn.0513-353x.2014-0999 |
| Jiang TT, Gao YH, Tong ZK. Selection of reference genes for quantitative real-time PCR in Lycoris[J]. Acta Hortic Sin, 2015, 42(6): 1129-1138. | |
| [19] |
马璐琳, 段青, 崔光芬, 等. 钝裂银莲花花色素合成相关基因RT-qPCR内参基因的筛选[J]. 园艺学报, 2021, 48(2): 377-388.
doi: 10.16420/j.issn.0513-353x.2020-0306 |
| Ma LL, Duan Q, Cui GF, et al. Selection and validation of reference genes for RT-qPCR analysis of the correlated genes in flower pigments biosynthesis pathway of Anemone obtusiloba[J]. Acta Hortic Sin, 2021, 48(2): 377-388. | |
| [20] | 李梦倩, 刘敏, 张蒙, 等. 大蒜体细胞胚发生mRNA及miRNA qPCR内参基因的筛选和验证[J]. 农业生物技术学报, 2021, 29(12): 2449-2464. |
| Li MQ, Liu M, Zhang M, et al. Identification and verification of somatic embryogenesis mRNA and miRNA qPCR reference genes in garlic(Allium sativum)[J]. J Agric Biotechnol, 2021, 29(12): 2449-2464. | |
| [21] |
Gutierrez L, Mauriat M, Guénin S, et al. The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction(RT-PCR)analysis in plants[J]. Plant Biotechnol J, 2008, 6(6): 609-618.
doi: 10.1111/j.1467-7652.2008.00346.x pmid: 18433420 |
| [22] |
Volkov RA, Panchuk II, Schöffl F. Heat-stress-dependency and developmental modulation of gene expression: the potential of house-keeping genes as internal standards in mRNA expression profiling using real-time RT-PCR[J]. J Exp Bot, 2003, 54(391): 2343-2349.
pmid: 14504302 |
| [23] |
Hu RB, Fan CM, Li HY, et al. Evaluation of putative reference genes for gene expression normalization in soybean by quantitative real-time RT-PCR[J]. BMC Mol Biol, 2009, 10: 93.
doi: 10.1186/1471-2199-10-93 pmid: 19785741 |
| [24] |
Die JV, Román B, Nadal S, et al. Evaluation of candidate reference genes for expression studies in Pisum sativum under different experimental conditions[J]. Planta, 2010, 232(1): 145-153.
doi: 10.1007/s00425-010-1158-1 URL |
| [25] |
Kozera B, Rapacz M. Reference genes in real-time PCR[J]. J Appl Genet, 2013, 54(4): 391-406.
pmid: 24078518 |
| [26] | 张玉芳, 赵丽娟, 曾幼玲. 基因表达研究中内参基因的选择与应用[J]. 植物生理学报, 2014, 50(8): 1119-1125. |
| Zhang YF, Zhao LJ, Zeng YL. Selection and application of reference genes for gene expression studies[J]. Plant Physiol J, 2014, 50(8): 1119-1125. | |
| [27] |
Deo RC, Bonanno JB, Sonenberg N, et al. Recognition of polyadenylate RNA by the poly(A)-binding protein[J]. Cell, 1999, 98(6): 835-845.
pmid: 10499800 |
| [28] |
Rocak S, Linder P. DEAD-box proteins: the driving forces behind RNA metabolism[J]. Nat Rev Mol Cell Biol, 2004, 5(3): 232-241.
doi: 10.1038/nrm1335 |
| [29] | 蔡敬, 孟小庆, 董婷婷, 等. DEAD-box解旋酶在植物非生物胁迫响应中的功能研究进展[J]. 生命科学, 2017, 29(5): 427-433. |
| Cai J, Meng XQ, Dong TT, et al. Progress of plant DEAD-box helicase in response to abiotic stress[J]. Chin Bull Life Sci, 2017, 29(5): 427-433. | |
| [30] | Vandesompele J, de Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes[J]. Genome Biol, 2002, 3(7): 467-470. |
| [31] |
Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets[J]. Cancer Res, 2004, 64(15): 5245-5250.
doi: 10.1158/0008-5472.CAN-04-0496 pmid: 15289330 |
| [32] |
Pfaffl MW, Tichopad A, Prgomet C, et al. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-based tool using pair-wise correlations[J]. Biotechnol Lett, 2004, 26(6): 509-515.
doi: 10.1023/B:BILE.0000019559.84305.47 URL |
| [33] | 吴建阳, 何冰, 杜玉洁, 等. 利用geNorm、NormFinder和BestKeeper软件进行内参基因稳定性分析的方法[J]. 现代农业科技, 2017(5): 278-281. |
| Wu JY, He B, Du YJ, et al. Analysis method of systematically evaluating stability of reference genes using geNorm, NormFinder and BestKeeper[J]. Mod Agric Sci Technol, 2017(5): 278-281. | |
| [34] |
Xie F, Xiao P, Chen D, et al. miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs[J]. Plant Mol Biol, 2012, 80(1):75-84.
doi: 10.1007/s11103-012-9885-2 URL |
| [35] | Ann M. Regulatory and functional interactions of plant growth regulators and plant glutathione S-transferases(GSTs)[J]. Vitam Horm, 2005, 72: 155-202. |
| [36] |
Agrawal GK, Rakwal R, Jwa NS, et al. Signalling molecules and blast pathogen attack activates rice OsPR1a and OsPR1b genes: a model illustrating components participating during defence/stress response[J]. Plant Physiol Biochem, 2001, 39(12): 1095-1103.
doi: 10.1016/S0981-9428(01)01333-X URL |
| [37] |
van Loon LC, Rep M, Pieterse CMJ. Significance of inducible defense-related proteins in infected plants[J]. Annu Rev Phytopathol, 2006, 44: 135-162.
pmid: 16602946 |
| [38] | 张亚琳, 张亚琨, 江春泉, 等. 人参锈腐病菌Ilyonectria robusta侵染人参根实时定量PCR内参基因的筛选与验证[J/OL]. 吉林农业大学学报, 2022. https://kns.cnki.net/kcms/detail/22.1100.S.20220331.1519.003.html. |
| Zhang YL, Zhang YK, Jiang CQ, et al. Screening and verification of internal reference genes in Panax ginseng roots infected by Ilyonectria robusta causing ginseng rusty rot by real-time quantitative PCR[J/OL]. J Jilin Agric Univ, 2022. https://kns.cnki.net/kcms/detail/22.1100.S.20220331.1519.003.html. | |
| [39] | 姚李祥, 潘春柳, 余丽莹, 等. 草果种子休眠解除过程中RT-qPCR内参基因筛选[J]. 中国中药杂志, 2021, 46(15): 3832-3837. |
| Yao LX, Pan CL, Yu LY, et al. Selection of RT-qPCR reference genes for Amomum tsaoko seeds during dormancy release[J]. China J Chin Mater Med, 2021, 46(15): 3832-3837. | |
| [40] | 赵艺蕊, 黄春颖, 王克涛, 等. 山核桃实时荧光定量PCR分析中内参基因的筛选与验证[J]. 果树学报, 2022, 39(1): 10-21. |
| Zhao YR, Huang CY, Wang KT, et al. Screening and verification of internal reference genes by real time quantitative PCR analysis in Carya cathayensis[J]. J Fruit Sci, 2022, 39(1): 10-21. | |
| [41] | 苏丹丹, 刘玉萍, 张雨, 等. 苦豆子实时荧光定量PCR内参基因筛选与验证[J]. 植物生理学报, 2022, 58(7): 1295-1306. |
| Su DD, Liu YP, Zhang Y, et al. Screening and validation of reference genes in Sophora alopecuroides for real-time quantitative PCR[J]. Plant Physiol J, 2022, 58(7): 1295-1306. | |
| [42] |
徐圆圆, 赵国春, 郝颖颖, 等. 无患子RT-qPCR内参基因的筛选与验证[J]. 生物技术通报, 2022, 38(10):80-89.
doi: 10.13560/j.cnki.biotech.bull.1985.2021-1616 |
| Xu YY, Zhao GC, Hao YY, et al. Reference genes selection and validation for RT-qPCR in Sapindus mukorossi[J]. Biotechnol Bull, 2022, 38(10):80-89. | |
| [43] |
庞强强, 李植良, 罗少波, 等. 高温胁迫下茄子RT-qPCR内参基因筛选及稳定性分析[J]. 园艺学报, 2017, 44(3): 475-486.
doi: 10.16420/j.issn.0513-353x.2016-0831 |
| Pang QQ, Li ZL, Luo SB, et al. Selection and stability analysis of reference gene for RT-qPCR in eggplant under high temperature stress[J]. Acta Hortic Sin, 2017, 44(3): 475-486. | |
| [44] |
Faccioli P, Ciceri GP, Provero P, et al. A combined strategy of “in silico” transcriptome analysis and web search engine optimization allows an agile identification of reference genes suitable for normalization in gene expression studies[J]. Plant Mol Biol, 2007, 63(5): 679-688.
doi: 10.1007/s11103-006-9116-9 URL |
| [45] |
Liu WG, Tang X, Qi XH, et al. The ubiquitin conjugating enzyme: an important ubiquitin transfer platform in ubiquitin-proteasome system[J]. Int J Mol Sci, 2020, 21(8): 2894.
doi: 10.3390/ijms21082894 URL |
| [46] |
Grumati P, Dikic I. Ubiquitin signaling and autophagy[J]. J Biol Chem, 2018, 293(15): 5404-5413.
doi: 10.1074/jbc.TM117.000117 pmid: 29187595 |
| [47] |
Nakamura N. Ubiquitin system[J]. International Journal of Molecular Sciences, 2018, 19(4):1080.
doi: 10.3390/ijms19041080 URL |
| [48] |
Hamey JJ, Wilkins MR. Methylation of elongation factor 1A: where, who, and why?[J]. Trends Biochem Sci, 2018, 43(3): 211-223.
doi: S0968-0004(18)30004-5 pmid: 29398204 |
| [49] |
Bevitori R, Oliveira MB, Grossi-de-Sá MF, et al. Selection of optimized candidate reference genes for RT-qPCR normalization in rice(Oryza sativa L.)during Magnaporthe oryzae infection and drought[J]. Genet Mol Res, 2014, 13(4): 9795-9805.
doi: 10.4238/2014.November.27.7 pmid: 25501189 |
| [50] | 代资举, 王艳, 王新涛, 等. 玉米粗缩病诱导下实时荧光定量PCR内参基因的选择[J]. 植物生理学报, 2019, 55(10): 1545-1553. |
| Dai ZJ, Wang Y, Wang XT, et al. Reference genes selection for real-time quantitative PCR under rough dwarf virus induced disease in maize[J]. Plant Physiol J, 2019, 55(10): 1545-1553. | |
| [51] | 唐枝娟, 刘秦, 肖晓蓉, 等. 白叶枯病菌侵染下的水稻内参基因稳定性[J]. 分子植物育种, 2017, 15(1): 300-306. |
| Tang ZJ, Liu Q, Xiao XR, et al. Selection of optimized candidate reference genes for RT-qPCR normalization in rice during Xanthomonas oryzae pv. oryzae infection[J]. Mol Plant Breed, 2017, 15(1): 300-306. | |
| [52] | 赵辉, 张春艳, 文艺, 等. 菜豆壳球孢侵染芝麻过程中内参基因的筛选[J]. 中国油料作物学报, 2017, 39(3): 393-398, 419. |
| Zhao H, Zhang CY, Wen Y, et al. Screening of reference genes in sesame during Macrophomina phaseolina infection[J]. Chin J Oil Crop Sci, 2017, 39(3): 393-398, 419. | |
| [53] | 姚全胜, 杨倩, 柳凤, 等. 芒果细菌性角斑病菌在侵染芒果叶片过程中内参基因筛选[J]. 分子植物育种, 2021, 19(18): 6088-6095. |
| Yao QS, Yang Q, Liu F, et al. Screening of reference genes in Xanthomonas citri pv. mangiferaeindicae during the infection of mango leaf[J]. Mol Plant Breed, 2021, 19(18): 6088-6095. | |
| [54] |
李恬静薇, 邹潇潇, 朱军, 等. 长茎葡萄蕨藻胁迫条件下RT-qPCR内参基因的筛选与验证[J]. 生物技术通报, 2021, 37(10): 266-276.
doi: 10.13560/j.cnki.biotech.bull.1985.2020-1585 |
| Li T, Zou XX, Zhu J, et al. Selection and validation of reference genes for quantitative real-time PCR in Caulerpa lentillifera under stress conditions[J]. Biotechnol Bull, 2021, 37(10): 266-276. | |
| [55] | 陈慧洁, 冯丽贞, 李慧敏, 等. 桉树焦枯病菌内参基因的筛选[J]. 森林与环境学报, 2018, 38(1): 98-103. |
| Chen HJ, Feng LZ, Li HM, et al. Screening and evaluation of reference genes of Calonectria pseudoreteaudii[J]. J For Environ, 2018, 38(1): 98-103. |
| [1] | 余慧, 王静, 梁昕昕, 辛亚平, 周军, 赵会君. 宁夏枸杞铁镉响应基因的筛选及其功能验证[J]. 生物技术通报, 2023, 39(7): 195-205. |
| [2] | 姚姿婷, 曹雪颖, 肖雪, 李瑞芳, 韦小妹, 邹承武, 朱桂宁. 火龙果溃疡病菌实时荧光定量PCR内参基因的筛选[J]. 生物技术通报, 2023, 39(5): 92-102. |
| [3] | 穆德添, 万凌云, 章瑶, 韦树根, 陆英, 付金娥, 田艺, 潘丽梅, 唐其. 钩藤管家基因筛选及生物碱合成相关基因的表达分析[J]. 生物技术通报, 2023, 39(2): 126-138. |
| [4] | 曹英芳, 赵新, 刘双, 李瑞环, 刘娜, 徐石勇, 高芳瑞, 马卉, 兰青阔, 檀建新, 王永. 抗除草剂大豆GE-J12实时荧光定量PCR检测方法的建立[J]. 生物技术通报, 2022, 38(7): 146-152. |
| [5] | 徐圆圆, 赵国春, 郝颖颖, 翁学煌, 陈仲, 贾黎明. 无患子RT-qPCR内参基因的筛选与验证[J]. 生物技术通报, 2022, 38(10): 80-89. |
| [6] | 陈建军, 赵怡迪, 曹香林. 脂多糖对鲤肠上皮细胞转录组模式的调控分析[J]. 生物技术通报, 2021, 37(8): 213-220. |
| [7] | 张静, 熊燕, 华永琳, 郭玉, 熊显荣, 字向东, 李键. 小鼠骨骼肌纤维类型定量PCR内参基因的筛选[J]. 生物技术通报, 2021, 37(2): 71-79. |
| [8] | 王欢禹, 常昊宛, 章崇祺, 金卫林, 魏芳. 五种检测嵌合抗原受体表达方法的比较[J]. 生物技术通报, 2021, 37(12): 265-273. |
| [9] | 李恬静薇, 邹潇潇, 朱军, 鲍时翔. 长茎葡萄蕨藻胁迫条件下RT-qPCR内参基因的筛选与验证[J]. 生物技术通报, 2021, 37(10): 266-276. |
| [10] | 潘沫晗, 陆添权, 田波. 滇牡丹种子实时荧光定量PCR分析中内参基因的筛选与验证[J]. 生物技术通报, 2020, 36(9): 218-226. |
| [11] | 庞鹏湘, 常燕楠, 尉瑞敏, 郜刚. 马铃薯StSRP1的克隆、表达及生物信息学分析[J]. 生物技术通报, 2019, 35(7): 10-16. |
| [12] | 孔春艳, 陈永坤, 王莎莎, 郝大海, 杨宇, 龚明. 小桐子低温胁迫下microRNA实时荧光定量PCR内参的筛选与比较[J]. 生物技术通报, 2019, 35(7): 25-32. |
| [13] | 朱锐, 叶雨情, 王雅欣, 杨晨茹, 王红伟, 孙晓晴, 张研, 李尚琪, 李炯棠. 鲤两种孕激素受体基因克隆、表达及比较分析[J]. 生物技术通报, 2019, 35(7): 46-53. |
| [14] | 王杰, 张阳, 秦澎, 何茂兰, 李津, 辜运富, 曾先富, 向泉桔. pH对香菇多糖含量及合成关键酶基因转录水平的影响[J]. 生物技术通报, 2019, 35(2): 39-45. |
| [15] | 张燕, 夏更寿, 赖志兵. 植物抗灰霉病菌分子机制的研究进展[J]. 生物技术通报, 2018, 34(2): 10-24. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||