








生物技术通报 ›› 2023, Vol. 39 ›› Issue (4): 81-92.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0771
收稿日期:2022-06-25
出版日期:2023-04-26
发布日期:2023-05-16
通讯作者:
任宇红,男,博士,教授,研究方向:生物催化、生物发酵过程;E-mail: yhren@ecust.edu.cn作者简介:韩惠,女,硕士研究生,研究方向:生物催化与酶工程;E-mail: hanhui16@126.com
HAN Hui( ), ZHANG Jian, REN Yu-hong(
), ZHANG Jian, REN Yu-hong( )
)
Received:2022-06-25
Published:2023-04-26
Online:2023-05-16
摘要:
2-氨基-1-(4-硝基苯基)-1,3-丙二醇俗称氯霉胺(ANP),由于具有两个手性中心,且结构中的O原子和N原子有良好的配位能力,具有广泛的应用价值。针对化学合成法存在生产成本高、原子经济性低、环保压力大等诸多缺点,旨在通过化学水解法与生物催化法相结合的方式,构建以对硝基-α-乙酰氨基-β-羟基苯丙酮(p-NAH)为底物合成(1R)-ANP的新途径。首先采用化学法水解p-NAH制备1-(4-硝基苯基)-2-氨基-3-羟基苯丙酮(AHNA),并筛选对水解产物具有催化活性的羰基还原酶,通过分子改造提高该酶的催化活性,并对突变体mut-V112Y的酶学性质进行研究;然后构建mut-V112Y与甲酸脱氢酶的双酶共表达及融合表达重组菌株,并对重组菌株的催化效率进行比较;最后优化催化反应条件,并进行制备反应。结果表明,化学法可水解p-NAH生成AHNA,筛选到的短链脱氢酶Lvchun可催化AHNA生成(1R)-ANP,通过对该酶进行定点突变获得了催化效率提高3.47倍的突变体mut-V112Y,其最适温度为30℃,最适pH为7.5,具有良好的温度和pH稳定性。成功构建了mut-V112Y和甲酸脱氢酶CbFDH的双酶共表达和融合表达重组菌株,通过比较发现共表达菌株mut-V112Y-CbFDH的催化效率最高。通过优化催化反应条件,最终可在最适条件下反应30 min催化50 mmol/L AHNA生成14.56 mmol/L(1R)-ANP,收率为29.12%。化学水解法与生物催化法相结合的方式可有效地催化p-NAH合成(1R)-ANP,该方法为合成光学纯的ANP提供了新途径。
韩惠, 张舰, 任宇红. 短链脱氢酶Lvchun的分子改造及其在氯霉胺合成中的应用[J]. 生物技术通报, 2023, 39(4): 81-92.
HAN Hui, ZHANG Jian, REN Yu-hong. Molecular Modification of the Short-chain Dehydrogenase Lvchun and Its Application in the Synthesis of Chloromycetin[J]. Biotechnology Bulletin, 2023, 39(4): 81-92.
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 大小 Size/bp |
|---|---|---|
| P1 | CGCGGATCCAAGGAGATATACATATGAAGATTGTCTTAGTTCTTTATGAT | 50 |
| P2 | CGGCTCGAGTTTCTTATCGTGTTTACCGTAAGCTTTAGTAACGTA | 45 |
| P3 | ACCACCACCACCACCACTGAAAGGAGATATACATATGGGCAGCA | 44 |
| P4 | GCTTTGTTAGCAGCCGGATCTCAGACCTGGCTGAAGCCG | 39 |
| P5 | GATCCGGCTGCTAACAAAGC | 20 |
| P6 | TCAGTGGTGGTGGTGGTGGT | 20 |
表1 共表达基因引物序列
Table 1 Primer sequences of co-expression genes
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 大小 Size/bp |
|---|---|---|
| P1 | CGCGGATCCAAGGAGATATACATATGAAGATTGTCTTAGTTCTTTATGAT | 50 |
| P2 | CGGCTCGAGTTTCTTATCGTGTTTACCGTAAGCTTTAGTAACGTA | 45 |
| P3 | ACCACCACCACCACCACTGAAAGGAGATATACATATGGGCAGCA | 44 |
| P4 | GCTTTGTTAGCAGCCGGATCTCAGACCTGGCTGAAGCCG | 39 |
| P5 | GATCCGGCTGCTAACAAAGC | 20 |
| P6 | TCAGTGGTGGTGGTGGTGGT | 20 |
| 短肽名称 Short peptide name | 氨基酸序列 Amino acid sequence | 大小 Size/aa |
|---|---|---|
| L1 | GGGGSGGGGS | 10 |
| L2 | GGGGSGGGGSGGGGS | 15 |
| L3 | EEEEKKKKEEEEKKKK | 15 |
| L4 | KAKLKEEEERKQREEEERIKRLEELAKRKEEERK | 34 |
| L5 | EEEEKKKQQEEEAERLRRIQEEMEKERKRREEDEERRRKEEEERRMKLEMEAKRKQEEEERKKREDDEKRKKK | 51 |
表2 连接短肽的氨基酸序列
Table 2 Amino acid sequences of linkers to short peptides
| 短肽名称 Short peptide name | 氨基酸序列 Amino acid sequence | 大小 Size/aa |
|---|---|---|
| L1 | GGGGSGGGGS | 10 |
| L2 | GGGGSGGGGSGGGGS | 15 |
| L3 | EEEEKKKKEEEEKKKK | 15 |
| L4 | KAKLKEEEERKQREEEERIKRLEELAKRKEEERK | 34 |
| L5 | EEEEKKKQQEEEAERLRRIQEEMEKERKRREEDEERRRKEEEERRMKLEMEAKRKQEEEERKKREDDEKRKKK | 51 |

图4 Lvchun与底物AHNA的分子对接 黄色表示辅酶NADH,蓝色表示氨基酸残基,绿色表示底物AHNA
Fig. 4 Molecular docking of Lvchun and substrate AHNA Yellow indicates coenzyme NADH, blue indicates amino acid residue, and green indicates substrate AHNA

图6 mut-V112Y在E. coli BL21(DE3)中的表达和纯化 M:标准蛋白分子量;1:粗酶液;2:穿出液;3:50 mmol/L咪唑洗脱液;4:200 mmol/L咪唑洗脱液;5:500 mmol/L咪唑洗脱液
Fig. 6 Expression and purification of mut-V112Y in E. coli BL21(DE3) M: Marker. 1: Crude enzyme solution. 2: Wear-off solution. 3: 50 mmol/L imidazole eluent. 4: 200 mmol/L imidazole eluent. 5: 500 mmol/L imidazole eluent
| 酶 Enzyme | 比活力Specific activity/(U·mg-1) | 米氏常数Km/(mmol·L-1) | 转换数kcat/(s-1) | 催化效率常数kcat/Km/(mmol·L-1·s-1) |
|---|---|---|---|---|
| Lvchun | 5.64 | 2.45 | 68.22 | 27.84 |
| mut-V112Y | 10.06 | 1.57 | 85.18 | 54.25 |
表3 Lvchun及mut-V112Y的酶活及动力学常数
Table 3 Specific activity and kinetic parameters of Lvchun and mut-V112Y
| 酶 Enzyme | 比活力Specific activity/(U·mg-1) | 米氏常数Km/(mmol·L-1) | 转换数kcat/(s-1) | 催化效率常数kcat/Km/(mmol·L-1·s-1) |
|---|---|---|---|---|
| Lvchun | 5.64 | 2.45 | 68.22 | 27.84 |
| mut-V112Y | 10.06 | 1.57 | 85.18 | 54.25 |
| 重组菌株名称 Name of recombinant strain | 羰基还原酶 mut-V112Y | 甲酸脱氢酶 CbFDH | 融合蛋白 Fusion proteins | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T | S | P | T | S | P | T | S | P | ||||
| mut-V112Y-CbFDH | + + + + | + + + + | - | + + + | + + + | - | - | - | - | |||
| CbFDH-mut-V112Y | + + + | + + + | - | + + | + + | - | - | - | - | |||
| F-L1-LY | - | - | - | - | - | - | + + + | + + | + + | |||
| F-L2-LY | - | - | - | - | - | - | + + + | + + | + + | |||
| F-L3-LY | - | - | - | - | - | - | + + | + + | - | |||
| F-L4-LY | - | - | - | - | - | - | + | + | - | |||
| F-L5-LY | - | - | - | - | - | - | + | + | - | |||
| LY-L1-F | - | - | - | - | - | - | + + + | + + | + + | |||
| LY-L2-F | - | - | - | - | - | - | + + + | + + | + + | |||
| LY-L3-F | - | - | - | - | - | - | + + + | + + + | + | |||
| LY-L4-F | - | - | - | - | - | - | + + + | + + + | + | |||
| LY-L5-F | - | - | - | - | - | - | + + + | + + + | + | |||
表4 重组蛋白在大肠杆菌中的表达情况
Table 4 Expressions of recombinant proteins in E. coli BL21(DE3)
| 重组菌株名称 Name of recombinant strain | 羰基还原酶 mut-V112Y | 甲酸脱氢酶 CbFDH | 融合蛋白 Fusion proteins | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T | S | P | T | S | P | T | S | P | ||||
| mut-V112Y-CbFDH | + + + + | + + + + | - | + + + | + + + | - | - | - | - | |||
| CbFDH-mut-V112Y | + + + | + + + | - | + + | + + | - | - | - | - | |||
| F-L1-LY | - | - | - | - | - | - | + + + | + + | + + | |||
| F-L2-LY | - | - | - | - | - | - | + + + | + + | + + | |||
| F-L3-LY | - | - | - | - | - | - | + + | + + | - | |||
| F-L4-LY | - | - | - | - | - | - | + | + | - | |||
| F-L5-LY | - | - | - | - | - | - | + | + | - | |||
| LY-L1-F | - | - | - | - | - | - | + + + | + + | + + | |||
| LY-L2-F | - | - | - | - | - | - | + + + | + + | + + | |||
| LY-L3-F | - | - | - | - | - | - | + + + | + + + | + | |||
| LY-L4-F | - | - | - | - | - | - | + + + | + + + | + | |||
| LY-L5-F | - | - | - | - | - | - | + + + | + + + | + | |||
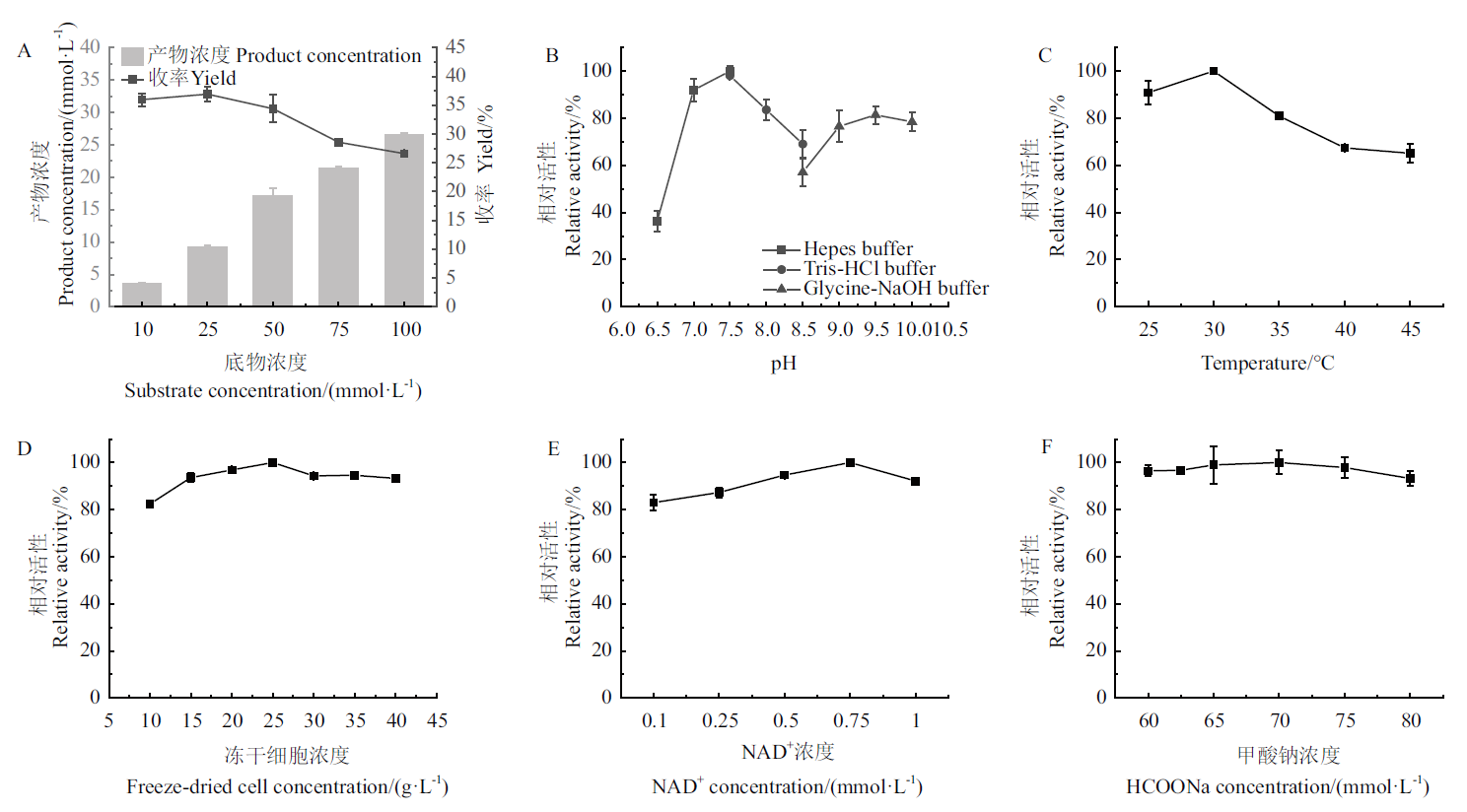
图9 反应条件的优化 A:底物耐受性;B:pH;C:温度;D:冻干细胞浓度;E:NAD+浓度;F:HCOONa浓度
Fig. 9 Optimization of reaction conditions A: Substrate tolerance; B: pH; C: temperature; D: concentration of lyophilized cell; E: concentration of NAD+; F: concentration of HCOONa
| [1] | 贾美荣, 杨康辉, 江余祺, 等. 手性氯霉胺及其衍生物的应用进展[J]. 中国药物化学杂志, 2010, 20(6): 543-549, 551. |
| Jia MR, Yang KH, Jiang YQ, et al. The development of chiral chloramphenicol base and its derivatives[J]. Chin J Med Chem, 2010, 20(6): 543-549, 551. | |
| [2] |
Ha WZ, Shan ZX. An economic, practical access to enantiopure 1, 1'-bi-2-naphthols: enantioselective complexation of threo-(1S, 2S)-N-benzyl-N, N-dimethyl[1, 3-dihydroxy-1-(4'-nitrophenyl)]-2-propylammonium chloride[J]. Tetrahedron Asymmetry, 2006, 17(5): 854-859.
doi: 10.1016/j.tetasy.2006.02.015 URL |
| [3] |
Jiang B, Si YG. The first highly enantioselective alkynylation of chloral: a practical and efficient pathway to chiral trichloromethyl propargyl alcohols[J]. Adv Synth Catal, 2004, 346(6): 669-674.
doi: 10.1002/(ISSN)1615-4169 URL |
| [4] |
Tang HY, Zhao GF, Zhou ZH, et al. Chiral tertiary amine/L-proline cocatalyzed enantioselective morita-baylis-Hillman(MBH)reaction[J]. Eur J Org Chem, 2008, 2008(1): 126-135.
doi: 10.1002/(ISSN)1099-0690 URL |
| [5] |
Feng XC, Qiu GF, Liang SC, et al. Efficient synthesis of chiral β-and γ-N-tosylaminoalcohols from 1-aryl-2-aminopropane-1, 3-diols[J]. Russ J Org Chem, 2006, 42(4): 496-500.
doi: 10.1134/S107042800604004X URL |
| [6] |
Madesclaire M, Coudert P, Zaitsev VP, et al. Regioselectivity of the interaction of(1S, 2S)-2-amino- 1-(4-nitrophenyl)-1, 3-propanediol with some symmetrical ketones[J]. Chem Heterocycl Compd, 2004, 40(10): 1310-1314.
doi: 10.1007/s10593-005-0066-y URL |
| [7] |
Hazra B, Pore V, Dey S, et al. Bile acid amides derived from chiral amino alcohols: novel antimicrobials and antifungals[J]. Bioorg Med Chem Lett, 2004, 14(3): 773-777.
pmid: 14741287 |
| [8] | 杨尚金, 冯珂, 朱毅, 等. 一种氯霉素的合成方法: CN102399160A[P]. 2012-04-04. |
| Yang SJ, Feng K, Zhu Y, et al. Method for synthesizing chloramphenicol: CN102399160A[P]. 2012-04-04. | |
| [9] | 杨尚金, 冯珂, 朱毅, 等. 一种由硝基甲烷合成氯霉素的方法: CN102399164B[P]. 1970-01-17. |
| Yang SJ, Feng K, Zhu Y, et al. Method for synthesizing chloramphenicol from nitromethane: CN102399164B[P]. 1970-01-17. | |
| [10] | 谢新开, 黄晓飞, 杜好勉. 一种制备氯霉素的方法: CN111662937B[P]. 2021-09-03. |
| Xie XK, Huang XF, Du HM. Method for preparing chloramphenicol: CN111662937B[P]. 2021-09-03. | |
| [11] | 谢新开, 黄晓飞, 张金鑫, 等. 一种氯霉素类化合物的制备方法: CN106566851B[P]. 2020-11-10. |
| Xie XK, Huang XF, Zhang JX, et al. Preparation method of chloramphenicol compounds: CN106566851B[P]. 2020-11-10. | |
| [12] |
Sato R, Amao Y. Can formate dehydrogenase from Candida boidinii catalytically reduce carbon dioxide, bicarbonate, or carbonate to formate?[J]. New J Chem, 2020, 44(28): 11922-11926.
doi: 10.1039/D0NJ01183E URL |
| [13] |
Li J, Feng JH, Chen X, et al. Structure-guided directed evolution of a carbonyl reductase enables the stereoselective synthesis of(2S, 3S)-2, 2-disubstituted-3-hydroxycyclopentanones via desymmetric reduction[J]. Org Lett, 2020, 22(9): 3444-3448.
doi: 10.1021/acs.orglett.0c00892 URL |
| [14] |
Deng J, Yao ZQ, Chen KL, et al. Towards the computational design and engineering of enzyme enantioselectivity: a case study by a carbonyl reductase from Gluconobacter oxydans[J]. J Biotechnol, 2016, 217: 31-40.
doi: 10.1016/j.jbiotec.2015.11.003 URL |
| [15] |
Crowe J, Masone BS, Ribbe J. One-step purification of recombinant proteins with the 6xHis tag and Ni-NTA resin[J]. Mol Biotechnol, 1995, 4(3): 247-258.
pmid: 8680931 |
| [16] |
Tang W, Chen LL, Deng J, et al. Structure-guided evolution of carbonyl reductase for efficient biosynthesis of ethyl(R)-2-hydroxy-4-phenylbutyrate[J]. Catal Sci Technol, 2020, 10(22): 7512-7522.
doi: 10.1039/D0CY01411G URL |
| [17] |
Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding[J]. Anal Biochem, 1976, 72: 248-254.
pmid: 942051 |
| [18] |
Wang Y, Li LX, Ma CQ, et al. Engineering of cofactor regeneration enhances(2S, 3S)-2, 3-butanediol production from diacetyl[J]. Sci Rep, 2013, 3: 2643.
doi: 10.1038/srep02643 pmid: 24025762 |
| [19] |
Lineweaver H, Burk D. The determination of enzyme dissociation constants[J]. J Am Chem Soc, 1934, 56(3): 658-666.
doi: 10.1021/ja01318a036 URL |
| [20] |
Zhan JR, Shou C, Zheng YC, et al. Discovery and engineering of bacterial(-)-isopiperitenol dehydrogenases to enhance(-)-menthol precursor biosynthesis[J]. Adv Synth Catal, 2021, 363(16): 3973-3982.
doi: 10.1002/adsc.v363.16 URL |
| [21] |
Moreira C, Ramos MJ, Fernandes PA. Glutamine synthetase drugability beyond its active site: exploring oligomerization interfaces and pockets[J]. Molecules, 2016, 21(8): 1028.
doi: 10.3390/molecules21081028 URL |
| [22] |
Koumanov A, Benach J, Atrian S, et al. The catalytic mechanism of Drosophila alcohol dehydrogenase: evidence for a proton relay modulated by the coupled ionization of the active site Lysine/Tyrosine pair and a NAD+ ribose OH switch[J]. Proteins, 2003, 51(2): 289-298.
doi: 10.1002/prot.v51:2 URL |
| [23] |
Eixelsberger T, Woodley JM, Nidetzky B, et al. Scale-up and intensification of(S)-1-(2-chlorophenyl)ethanol bioproduction: economic evaluation of whole cell-catalyzed reduction of o-chloroacetophenone[J]. Biotechnol Bioeng, 2013, 110(8): 2311-2315.
doi: 10.1002/bit.24896 pmid: 23475609 |
| [24] |
He L, Ye WJ, Xie YY, et al. Efficient biocatalytic synthesis of(R)-2-chloro-1-(3, 4-difluorophenyl)ethanol by the short-chain dehydrogenase PpKR8 from Paraburkholderia phymatum STM815[J]. Org Process Res Dev, 2022, 26(2): 278-287.
doi: 10.1021/acs.oprd.1c00189 URL |
| [25] |
Kizaki N, Yasohara Y, Hasegawa J, et al. Synthesis of optically pure ethyl(S)-4-chloro-3-hydroxybutanoate by Escherichia coli transformant cells coexpressing the carbonyl reductase and glucose dehy-drogenase genes[J]. Appl Microbiol Biotechnol, 2001, 55(5): 590-595.
pmid: 11414326 |
| [26] |
Teng Y, Gu CL, Chen ZH, et al. Advances and applications of chiral resolution in pharmaceutical field[J]. Chirality, 2022, 34(8): 1094-1119.
doi: 10.1002/chir.23453 pmid: 35676772 |
| [1] | 朱秋雨, 段绪果. L-天冬氨酸-α-脱羧酶的重组表达、定点突变及高通量检测方法的建立[J]. 生物技术通报, 2022, 38(5): 269-278. |
| [2] | 付雅丽, 彭万里, 林双君, 邓子新, 梁如冰. 香茅醇假单胞菌SJTE-3的短链脱氢酶SDR-X1的克隆及酶性质测定[J]. 生物技术通报, 2022, 38(3): 121-129. |
| [3] | 田庚, 高伟强, 陈晓波, 张春晓. 地衣芽孢杆菌KD-1β-甘露聚糖酶定点突变提高酶活性及稳定性[J]. 生物技术通报, 2021, 37(10): 100-109. |
| [4] | 仲建锋, 李兴奎, 徐重新, 张霄, 刘贤金. Cry1B抗独特型单链抗体的定点突变及生物活性分析[J]. 生物技术通报, 2021, 37(10): 186-195. |
| [5] | 孙熙麟, 蒋振彦, 刘志屹, 戴璐, 孙非, 黄伟. 氨基酸定点突变提高灵芝蛋白LZ-8热稳定性的研究[J]. 生物技术通报, 2020, 36(1): 23-28. |
| [6] | 邱锦, 黄火清, 姚斌, 罗会颖. 解淀粉芽孢杆菌淀粉酶催化活力改良及其在枯草芽孢杆菌中的高效表达[J]. 生物技术通报, 2019, 35(9): 134-143. |
| [7] | 徐林娜, 胡孟可, 童文艳, 李芬. 烟草NtTkr尾部点突变对卷曲螺旋结构及与靶蛋白相互作用的影响[J]. 生物技术通报, 2019, 35(5): 64-69. |
| [8] | 王柳月, 李慧美, 马梦琪, 梁明星, 贺如阳, 陈华波. 利用旁侧引物提高重叠延伸PCR定点突变效率[J]. 生物技术通报, 2019, 35(12): 196-202. |
| [9] | 华晨, 李新新, 涂涛, 杨虹, 罗会颖, 陈家明, 姚斌, 柏映国, 彭书传. 基于酶热稳定性系统计算的乳酸氧化酶热稳定性改造[J]. 生物技术通报, 2018, 34(8): 144-150. |
| [10] | 陈少威, 吴程, 苏月华, 蔡斌斌, 谢盼盼, 杨梅. 苏云金芽胞杆菌aiiA的5'端侧翼序列的克隆与功能鉴定[J]. 生物技术通报, 2018, 34(11): 136-143. |
| [11] | 秦海彬, 熊涛, 张博, 牛坤. α-酮戊二酸半醛脱氢酶的定点突变及酶学性质变化 [J]. 生物技术通报, 2017, 33(8): 180-185. |
| [12] | 曾静, 郭建军, 袁林, 杨罡, 陈俊. 极端嗜热α-淀粉酶ApkA的高温活性和热稳定性的优化研究[J]. 生物技术通报, 2017, 33(8): 192-198. |
| [13] | 刘松,陆信曜,周景文,堵国成,陈坚. 脂肪氧合酶结构、分子改造与发酵研究进展[J]. 生物技术通报, 2015, 31(12): 34-41. |
| [14] | 邬志杰, 吴更, 唐鸿志, 许平. 恶臭假单胞菌HspB的单晶培养及结晶条件优化[J]. 生物技术通报, 2015, 31(11): 236-242. |
| [15] | 庞浩, 陈燕, 吴倩倩, 刘春宇, 郭媛, 林丽华, 黄日波. 地衣芽孢杆菌(Bacillus licheniformis)外切葡聚糖酶CelB基因的发掘及功能鉴定[J]. 生物技术通报, 2013, 0(9): 151-157. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||