








生物技术通报 ›› 2023, Vol. 39 ›› Issue (4): 71-80.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0641
收稿日期:2022-05-24
出版日期:2023-04-26
发布日期:2023-05-16
通讯作者:
郑菲,女,博士,讲师,研究方向:微生物代谢与酶工程;E-mail: zhengfei0718@bjfu.edu.cn作者简介:杨俊钊,女,硕士研究生,研究方向:资源环境微生物学;E-mail: YJZbio@bjfu.edu.cn
YANG Jun-zhao( ), ZHANG Xin-rui, ZHAO Guo-zhu, ZHENG Fei(
), ZHANG Xin-rui, ZHAO Guo-zhu, ZHENG Fei( )
)
Received:2022-05-24
Published:2023-04-26
Online:2023-05-16
摘要:
纤维素酶能够将纤维素转化为可发酵的糖类,为丰富纤维素酶的序列与结构资源、揭示纤维素酶结构与功能之间的关系,本研究对两个新型GH5家族多结构域内切纤维素酶TlCel5和ReCel5进行了克隆表达和酶学性质测定,并对其结构域开展了突变研究。序列和结构分析显示,Tlcel5和Recel5分别编码了655个和632个氨基酸,理论分子量分别为68.3 kD和65.9 kD,均包含了CBM1区、CD区、CBMX2区和一个未知结构域,这与以往报道的多数单一结构域或双结构域纤维素酶显著不同。为了解附加结构域对酶功能的影响效果,以ReCel5为研究对象,分别构建了N端CBM1结构域的截断突变体TM1,和C端未知结构域的截断突变体TM2。酶学性质分析显示,TlCel5和ReCel5的最适作用pH和最适作用温度分别为pH 3.0、50℃和pH 4.0、70℃,在50℃和70℃下能够保持良好的稳定性,并且对多种类型的纤维素类底物、半纤维素类底物表现出水解能力。虽然突变体TM1和TM2的酶学性质较野生型没有发生显著变化,但其对羧甲基纤维素钠、大麦葡聚糖、地衣多糖的水解比活值降低了23%-68%,由此说明,在多结构域酶中,附加结构域与酶的水解能力之间存在密切关系。
杨俊钊, 张新蕊, 赵国柱, 郑菲. 新型GH5家族多结构域纤维素酶的结构与功能研究[J]. 生物技术通报, 2023, 39(4): 71-80.
YANG Jun-zhao, ZHANG Xin-rui, ZHAO Guo-zhu, ZHENG Fei. Structure and Function Analysis of Novel GH5 Multi-domain Cellulase[J]. Biotechnology Bulletin, 2023, 39(4): 71-80.
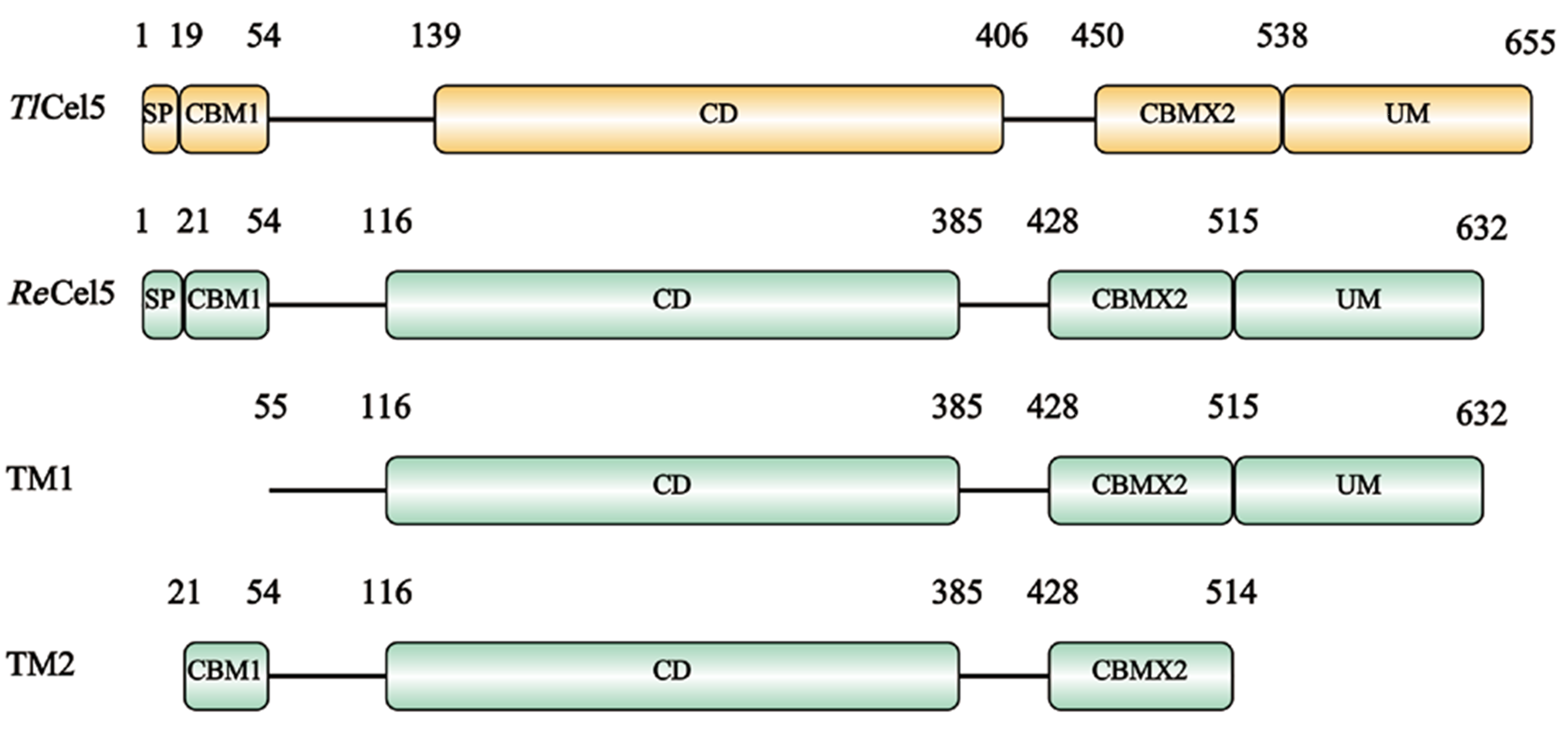
图1 TlCel5、ReCel5、TM1和TM2编码蛋白质结构示意图 SP:信号肽;CBM:碳水化合物结合模块;CD:催化结构域;UM:未知结构域
Fig. 1 Schematic diagram of protein structure encoded by TlCel5, ReCel5, TM1 and TM2 SP: Signal peptide. CBM: Carbohydrate binding module. CD: Catalysis domain. UM: Unknown module

图2 GH5家族内切纤维素酶催化结构域氨基酸序列对比图 保守残基由向下箭头所标记。比对序列的菌株来源及登录号分别为:H. jecorina(AEJ36301.1), R. emersonii CBS 393.64(XP_013323622.1), Aspergillus neoniger CBS 115656(XP_025478804.1), Thermoascus aurantiacus(AAL88714.2), Stegonsporium opalus(ARO48344.1), Ganoderma lucidum(QDK64599.1), Trichoderma reesei(P07982.1), and Gloeophyllum trabeum ATCC 11539(XP_007867902.1)
Fig. 2 Amino acids sequence alignment in the catalytic domain of GH5 family endoglucanases The conservative residues are labeled in the arrow below the alignment. The strain source and GenBank number of the alignment sequence are H. jecorina(AEJ36301.1), R. emersonii CBS 393.64(XP_013323622.1), A. neoniger CBS 115656(XP_025478804.1), T. aurantiacus(AAL88714.2), S. opalus(ARO48344.1), G. lucidum(QDK64599.1), T. reesei(P07982.1), and G. trabeum ATCC 11539(XP_007867902.1)

图3 野生型TlCel5、ReCel5和突变体TM1、TM2的SDS-PAGE分析 M:蛋白分子质量标准; A:野生型TlCel5、ReCel5的表达(1:ReCel5纯化蛋白;2:TlCel5纯化蛋白); B:突变体TM1、TM2的表达(3:突变体TM1纯化蛋白;4:突变体TM2纯化蛋白)
Fig. 3 SDS-PAGE analysis of wild-type TlCel5, ReCel5 and mutant TM1, TM2 M: Protein molecular weight standard. A: Expression of wile-type TlCel5、ReCel5(1: purified protein of ReCel5; 2: purified protein of TlCel5). B: Expressions of mutants TM1 and TM2(3: purified protein of TM1; 4: purified protein of TM2)

图4 野生型TlCel5、ReCel5及突变体TM1、TM2的酶学性质图 A:最适pH;B:pH稳定性;C:最适温度;D:50℃下温度稳定性;E:60℃下温度稳定性;F:70℃下温度稳定性
Fig. 4 Enzymatic properties of wild-type TlCel5, ReCel5 and mutant TM1, TM2 A: pH-activity profile. B: pH stability. C: Temperature-activity profile. D: Thermostability at 50℃. E: Thermostability at 60℃. F: Thermostability at 70℃
| Enzyme | Vmax/(mmol·min-1·mg-1) | Km/(mg·mL-1) | kcat/(s-1) | kcat/Km(mL·mg-1·s-1) |
|---|---|---|---|---|
| TlCel5 | 217.0±20.0 | 7.5±1.0 | 247.0±22.0 | 32.8±1.5 |
| ReCel5 | 56.0±0.9 | 8.6±0.2 | 61.4±0.9 | 7.1±0.1 |
| TM1 | 68.0±0.5 | 8.4±0.1 | 70.7±0.6 | 8.4±0.1 |
| TM2 | 76.9±0.7 | 8.4±0.1 | 68.1±0.6 | 8.1±0.1 |
表1 野生型TlCel5、ReCel5及突变体TM1、TM2的酶促反应动力学参数
Table 1 Kinetics parameters of wild-type TlCel5, ReCel5 and mutants TM1, TM2
| Enzyme | Vmax/(mmol·min-1·mg-1) | Km/(mg·mL-1) | kcat/(s-1) | kcat/Km(mL·mg-1·s-1) |
|---|---|---|---|---|
| TlCel5 | 217.0±20.0 | 7.5±1.0 | 247.0±22.0 | 32.8±1.5 |
| ReCel5 | 56.0±0.9 | 8.6±0.2 | 61.4±0.9 | 7.1±0.1 |
| TM1 | 68.0±0.5 | 8.4±0.1 | 70.7±0.6 | 8.4±0.1 |
| TM2 | 76.9±0.7 | 8.4±0.1 | 68.1±0.6 | 8.1±0.1 |
| Name | Optimal pH | Optimal temperature /℃ | Source | Reference |
|---|---|---|---|---|
| TlCel5 | 3.0 | 50 | T. leycettanus | This study |
| ReCel5 | 4.0 | 70 | R. emersonii | This study |
| GlCel5A | 3.0-4.0 | 60 | Ganoderma lucidum | [ |
| TaCel5A | 6.0 | 50 | Thermoascus aurantiacusIFO9748 | [ |
| Epi3 | 6.5-7.0 | 50 | Epidinium caudatum | [ |
| PdCel5C | 4.8 | 40-50 | Penicillium decumbens 114-2 | [ |
| BaCel5 | 5.0 | 50 | Bispora antennata | [ |
| SoCel5 | 5.0 | 60 | Stegonsporium opalus | [ |
表2 GH5家族真菌源内切纤维素酶最适条件表
Table 2 Optimal conditions for endoglucanases of GH5 family fungi
| Name | Optimal pH | Optimal temperature /℃ | Source | Reference |
|---|---|---|---|---|
| TlCel5 | 3.0 | 50 | T. leycettanus | This study |
| ReCel5 | 4.0 | 70 | R. emersonii | This study |
| GlCel5A | 3.0-4.0 | 60 | Ganoderma lucidum | [ |
| TaCel5A | 6.0 | 50 | Thermoascus aurantiacusIFO9748 | [ |
| Epi3 | 6.5-7.0 | 50 | Epidinium caudatum | [ |
| PdCel5C | 4.8 | 40-50 | Penicillium decumbens 114-2 | [ |
| BaCel5 | 5.0 | 50 | Bispora antennata | [ |
| SoCel5 | 5.0 | 60 | Stegonsporium opalus | [ |

图6 纤维素酶CBM1序列比对 比对序列的纤维素酶名称、来源菌株及登录号分别为:ReCel5(R.emersonii CBS 393.64, XP_013323622.1), TrCel7A(T. reesei, CAH10320.1), TrCel7B(T. reesei, AAA34212.1), PdCel5C(P. decumbens, JQ319040.1), TrCel5C(T. reesei, AAA34213.1), ApCel5A(Aureobasidium pullulans, AEM23896.1)
Fig. 6 Sequence alignment of CBM1 domain of cellulose The cellulase name, source strain and GenBank number of the alignment sequence are ReCel5(R. emersonii CBS 393.64, XP_013323622.1), TrCel7A(T. reesei, CAH10320.1), TrCel7B(T. reesei, AAA34212.1), PdCel5C(P. decumbens, JQ319040.1), TrCel5C(T. reesei, AAA34213.1), ApCel5A(Aureobasidium pullulans, AEM23896.1)
| [1] | 赵琪, 李亚兰, 陈子欣, 等. 纤维素酶应用研究的最新进展[J]. 广州化工, 2014, 42(6): 21-23. |
| Zhao Q, Li YL, Chen ZX, et al. The recent application progress cellulase[J]. Guangzhou Chem Ind, 2014, 42(6): 21-23. | |
| [2] |
Xin DL, Blossom BM, Lu XY, et al. Improving cellulases hydrolytic action: an expanded role for electron donors of lytic polysaccharide monooxygenases in cellulose saccharification[J]. Bioresour Technol, 2022, 346: 126662.
doi: 10.1016/j.biortech.2021.126662 URL |
| [3] |
Sidar A, Albuquerque ED, Voshol GP, et al. Carbohydrate binding modules: diversity of domain architecture in amylases and cellulases from filamentous microorganisms[J]. Front Bioeng Biotechnol, 2020, 8: 871.
doi: 10.3389/fbioe.2020.00871 URL |
| [4] |
Hu YM, Li HN, Ran QP, et al. Effect of carbohydrate binding modules alterations on catalytic activity of glycoside hydrolase family 6 exoglucanase from Chaetomium thermophilum to cellulose[J]. Int J Biol Macromol, 2021, 191: 222-229.
doi: 10.1016/j.ijbiomac.2021.09.002 URL |
| [5] |
Shibata N, Suetsugu M, Kakeshita H, et al. A novel GH10 xylanase from Penicillium sp. accelerates saccharification of alkaline-pretreated bagasse by an enzyme from recombinant Trichoderma reesei expressing Aspergillus β-glucosidase[J]. Biotechnol Biofuels, 2017, 10: 278.
doi: 10.1186/s13068-017-0970-2 pmid: 29201142 |
| [6] |
Thongekkaew J, Ikeda H, Masaki K, et al. Fusion of cellulose binding domain from Trichoderma reesei CBHI to Cryptococcus sp. S-2 cellulase enhances its binding affinity and its cellulolytic activity to insoluble cellulosic substrates[J]. Enzyme Microb Technol, 2013, 52(4/5): 241-246.
doi: 10.1016/j.enzmictec.2013.02.002 URL |
| [7] | 孔海洋, 蒋肖, 王苑, 等. β-甘露聚糖酶Man5A和木聚糖酶Tlxyn11B的融合表达[J]. 生物工程学报, 2020, 36(9): 1849-1858. |
| Kong HY, Jiang X, Wang Y, et al. Fusion expression of β-mannanase Man5A and xylanase Tlxyn11B in Pichia pastoris[J]. Chin J Biotechnol, 2020, 36(9): 1849-1858. | |
| [8] |
Liu GZ, Li Q, Shang N, et al. Functional and structural analyses of a 1, 4-β-endoglucanase from Ganoderma lucidum[J]. Enzyme Microb Technol, 2016, 86: 67-74.
doi: 10.1016/j.enzmictec.2016.01.013 URL |
| [9] |
Hong J, Tamaki H, Yamamoto K, et al. Cloning of a gene encoding a thermo-stable endo-beta-1, 4-glucanase from Thermoascus aurantiacus and its expression in yeast[J]. Biotechnol Lett, 2003, 25(8): 657-661.
doi: 10.1023/A:1023072311980 URL |
| [10] |
Takenak A, D’Silva CG, Kudo H, et al. Molecular cloning, expression, and characterization of an endo-beta-1, 4-glucanase cDNA from Epidinium caudatum1[J]. J Gen Appl Microbiol, 1999, 45(2): 57-61.
pmid: 12501388 |
| [11] |
Liu GD, Qin YQ, Hu YB, et al. An endo-1, 4-β-glucanase PdCel5C from cellulolytic fungus Penicillium decumbens with distinctive domain composition and hydrolysis product profile[J]. Enzyme Microb Technol, 2013, 52(3): 190-195.
doi: 10.1016/j.enzmictec.2012.12.009 URL |
| [12] |
Zheng F, Huang HQ, Wang XY, et al. Improvement of the catalytic performance of a Bispora antennata cellulase by replacing the N-terminal semi-barrel structure[J]. Bioresour Technol, 2016, 218: 279-285.
doi: 10.1016/j.biortech.2016.06.094 URL |
| [13] | Zheng F, Vermaas JV, Zheng J, et al. Activity and thermostability of GH5 endoglucanase chimeras from mesophilic and thermophilic parents[J]. Appl Environ Microbiol, 2019, 85(5): e02079-e02018. |
| [14] |
Inoue H, Kishishita S, Kumagai A, et al. Contribution of a family 1 carbohydrate-binding module in thermostable glycoside hydrolase 10 xylanase from Talaromyces cellulolyticus toward synergistic enzymatic hydrolysis of lignocellulose[J]. Biotechnol Biofuels, 2015, 8: 77.
doi: 10.1186/s13068-015-0259-2 URL |
| [15] |
Várnai A, Siika-Aho M, Viikari L. Carbohydrate-binding modules(CBMs)revisited: reduced amount of water counterbalances the need for CBMs[J]. Biotechnol Biofuels, 2013, 6(1): 30.
doi: 10.1186/1754-6834-6-30 pmid: 23442543 |
| [16] |
Ståhlberg J, Johansson G, Pettersson G. A binding-site-deficient, catalytically active, core protein of endoglucanase III from the culture filtrate of Trichoderma reesei[J]. Eur J Biochem, 1988, 173(1): 179-183.
pmid: 3356188 |
| [17] |
Kraulis J, Clore GM, Nilges M, et al. Determination of the three-dimensional solution structure of the C-terminal domain of cellobiohydrolase I from Trichoderma reesei. A study using nuclear magnetic resonance and hybrid distance geometry-dynamical simulated annealing[J]. Biochemistry, 1989, 28(18): 7241-7257.
doi: 10.1021/bi00444a016 pmid: 2554967 |
| [18] |
Hervé C, Rogowski A, Blake AW, et al. Carbohydrate-binding modules promote the enzymatic deconstruction of intact plant cell walls by targeting and proximity effects[J]. Proc Natl Acad Sci USA, 2010, 107(34): 15293-15298.
doi: 10.1073/pnas.1005732107 pmid: 20696902 |
| [19] |
Bolam DN, Ciruela A, Mcqueen-Mason S, et al. Pseudomonas cellulose-binding domains mediate their effects by increasing enzyme substrate proximity[J]. Biochem J, 1998, 331(Pt 3): 775-781.
doi: 10.1042/bj3310775 URL |
| [20] |
Aich S, Datta S. Engineering of a highly thermostable endoglucanase from the GH7 family of Bipolaris sorokiniana for higher catalytic efficiency[J]. Appl Microbiol Biotechnol, 2020, 104(9): 3935-3945.
doi: 10.1007/s00253-020-10515-0 |
| [21] |
Linder M, Lindeberg G, Reinikainen T, et al. The difference in affinity between two fungal cellulose-binding domains is dominated by a single amino acid substitution[J]. FEBS Lett, 1995, 372(1): 96-98.
pmid: 7556652 |
| [22] |
Arola S, Linder MB. Binding of cellulose binding modules reveal differences between cellulose substrates[J]. Sci Rep, 2016, 6: 35358.
doi: 10.1038/srep35358 pmid: 27748440 |
| [23] |
Nishijima H, Nozaki K, Mizuno M, et al. Extra tyrosine in the carbohydrate-binding module of Irpex lacteus Xyn10B enhances its cellulose-binding ability[J]. Biosci Biotechnol Biochem, 2015, 79(5): 738-746.
doi: 10.1080/09168451.2014.996203 URL |
| [1] | 赵赛赛, 张小丹, 贾晓妍, 陶大炜, 刘可玉, 宁喜斌. 高产硝酸盐还原酶Staphylococcus simulans ZSJ6的复合诱变选育及其酶学性质研究[J]. 生物技术通报, 2023, 39(4): 103-113. |
| [2] | 王雨辰, 丁尊丹, 关菲菲, 田健, 刘国安, 伍宁丰. 耐热漆酶ba4基因鉴定与酶学性质分析[J]. 生物技术通报, 2022, 38(8): 252-260. |
| [3] | 毛国涛, 王杰, 王凯, 王方园, 曹乐言, 张宏森, 宋安东. 水生栖热菌漆酶TaLac的性质分析及对孔雀石绿染料的脱除[J]. 生物技术通报, 2022, 38(4): 261-268. |
| [4] | 常晴, 束月蓉, 王文韬, 蒋昊, 延泉德, 钱政, 高雪纯, 吴金鸿, 张勇. 来自Yeosuana marina sp. JLT21内切型海藻酸裂解酶的异源表达及酶学表征[J]. 生物技术通报, 2022, 38(2): 123-131. |
| [5] | 王小桃, 邹杭, 吴怡, 向省维, 吕华, 刘超兰, 林家富, 王欣荣, 褚以文, 宋涛. Paraglaciecola hydrolytica中新型β-琼胶酶Aga2的异源表达及酶学性质分析[J]. 生物技术通报, 2022, 38(11): 258-268. |
| [6] | 岑潇龙, 雷曦, 马诗云, 陈倩茹, 黄遵锡, 周峻沛, 张蕊. 金黄色葡萄球菌透明质酸裂解酶HylS的异源表达与特性研究[J]. 生物技术通报, 2022, 38(1): 157-167. |
| [7] | 田嘉慧, 封佳丽, 卢俊桦, 毛林静, 胡著然, 王莹, 楚杰. 一色齿毛菌漆酶LacT-1的分离纯化与性质研究[J]. 生物技术通报, 2021, 37(8): 186-194. |
| [8] | 张瑶心, 王亮节, 郑文, 徐汉琴, 郑恋, 钟静. 产几丁质酶的无色杆菌ZWW8的发酵产酶及酶学性质研究[J]. 生物技术通报, 2021, 37(4): 96-106. |
| [9] | 刘珊, 叶伟, 朱牧孜, 李赛妮, 邓张双, 章卫民. 一种新型酰基转移酶GPAT的克隆、表达与酶学性质研究[J]. 生物技术通报, 2021, 37(11): 257-266. |
| [10] | 赵海燕, 宋晨斌, 刘正亚, 马兴荣, 尚会会, 李安华, 关现军, 王建设. 来源于Laceyella sp.的α-淀粉酶基因克隆、重组表达及酶学性质研究[J]. 生物技术通报, 2020, 36(8): 23-33. |
| [11] | 王惠兰, 吴金勇, 陈祥松, 袁丽霞, 朱薇薇, 姚建铭. N-乙酰神经氨酸醛缩酶的固定化及固定化酶性质研究[J]. 生物技术通报, 2020, 36(6): 165-173. |
| [12] | 朱彩林, 吕祥, 夏小乐. 盖子区域氨基酸的定点突变对T1脂肪酶酶学性质的影响[J]. 生物技术通报, 2020, 36(11): 94-102. |
| [13] | 张庆芳, 逄飞, 于爽, 肖景惠, 窦少华, 迟乃玉. 海洋高产尿酸氧化酶菌株筛选鉴定及酶学性质研究[J]. 生物技术通报, 2019, 35(7): 61-69. |
| [14] | 董聪, 高庆华, 王玥, 罗同阳. 基于密码子优化的FAD依赖葡萄糖脱氢酶在毕赤酵母中的高效表达及酶学性质[J]. 生物技术通报, 2019, 35(7): 114-120. |
| [15] | 徐珊 ,李任强 ,郑振华 ,张云 ,孙爱君 ,胡云峰. 红树林微生物DH-2胞外蛋白酶的性质及产酶条件优化[J]. 生物技术通报, 2018, 34(6): 120-127. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||