








生物技术通报 ›› 2023, Vol. 39 ›› Issue (4): 93-102.doi: 10.13560/j.cnki.biotech.bull.1985.2022-1384
收稿日期:2022-11-09
出版日期:2023-04-26
发布日期:2023-05-16
通讯作者:
杨冬同为本文通讯作者作者简介:杨冬,男,高级工程师,研究方向:检验检测;E-mail: 2012855620@qq.com
基金资助:Received:2022-11-09
Published:2023-04-26
Online:2023-05-16
摘要:
为丰富生物降解黄曲霉毒素B1(AFB1)资源,研究已筛选到的枯草芽孢杆菌WTX1降解AFB1的胞外酶特性及其降解位点。通过降解与AFB1毒性位点结构相似的3种化合物,采用液相色谱串联三重四级杆质谱(UPLC-MS/MS)检测降解液。经层析分离,降解AFB1胞外酶主要有Ea、Eb两种。最适降解条件:降解酶Ea 降解温度为50℃,pH 5,降解率高达88.4%;降解酶Eb降解温度为37℃,pH 5,降解率为73.4%;Ea酶的作用位点为AFB1的呋喃环双键、香兰素内脂环、戊烯酮环结构,Eb酶的作用位点为AFB1的香兰素内脂环、戊烯酮环结构。酶组降解途径为通过水解作用,加氢及连续脱去-CO基团。菌株WTX1胞外酶通过水解作用破坏AFB1毒性位点,起到降解作用,该酶具有很好的抗逆性,温度和pH适应性适合工业化加工的苛刻条件。
杨冬, 唐璎. 枯草芽孢杆菌WTX1胞外酶降解AFB1酶学特性及降解位点分析[J]. 生物技术通报, 2023, 39(4): 93-102.
YANG Dong, TANG Ying. Enzymatic Characterization and Degradation Sites of AFB1 Degradation by the Extracellular Enzyme of Bacillus subtilis Strain WTX1[J]. Biotechnology Bulletin, 2023, 39(4): 93-102.
| 序号 Serial No. | 目标物 Target | 保留时间 Retention time/min | 母离子 Parent ion/(m/z) | 子离子 Daughter ion/(m/z) | 去簇电压 Declustering voltage/DP | 碰撞能量 Crash energy/CE |
|---|---|---|---|---|---|---|
| 1 | AFB1 | 4.71 | 313.0 | 285.1* | 140 | 31 |
| 240.9 | 49 | |||||
| 2 | 香兰醛/ Vanillin | 3.13 | 151.1 | 131.2* 81.1 | 120 | 32 41 |
| 3 | 5-羟甲基糠醛/5-(Hydroxymethyl)furfural | 3.78 | 126.1 | 97.3* 41.2 | 120 | 35 49 |
| 4 | 1-戊烯-3-酮/1-Penten-3-one | 2.85 | 56.1 | 27.2 57.1* | 120 | 30 47 |
表1 质谱检测条件
Table 1 Mass detection condition
| 序号 Serial No. | 目标物 Target | 保留时间 Retention time/min | 母离子 Parent ion/(m/z) | 子离子 Daughter ion/(m/z) | 去簇电压 Declustering voltage/DP | 碰撞能量 Crash energy/CE |
|---|---|---|---|---|---|---|
| 1 | AFB1 | 4.71 | 313.0 | 285.1* | 140 | 31 |
| 240.9 | 49 | |||||
| 2 | 香兰醛/ Vanillin | 3.13 | 151.1 | 131.2* 81.1 | 120 | 32 41 |
| 3 | 5-羟甲基糠醛/5-(Hydroxymethyl)furfural | 3.78 | 126.1 | 97.3* 41.2 | 120 | 35 49 |
| 4 | 1-戊烯-3-酮/1-Penten-3-one | 2.85 | 56.1 | 27.2 57.1* | 120 | 30 47 |
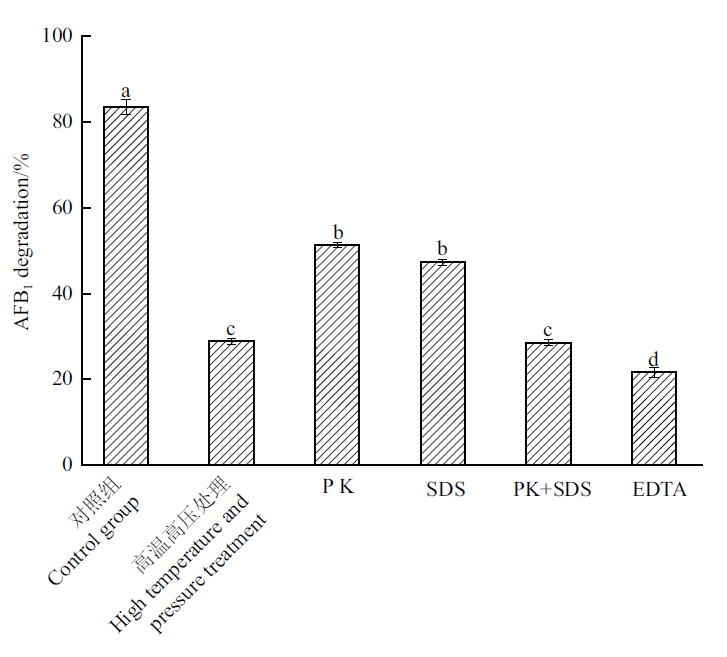
图1 不同变性处理对WTX1发酵上清液降解率影响 不同小写字母表示不同处理间显著性差异(P<0.05)。下同
Fig. 1 Effects of different denaturation treatments on the degradation rates of WTX1 strain fermentation supernatant Different lowercase letters indicate significant differences between different treatments(P < 0.05). The same below
| 酶 Enzyme | 香兰醛 Vanillin/% | 5-羟甲基糠醛 5-(Hydroxymethyl)furfural/% | 1-戊烯-3-酮 1-Penten-3-one/% |
|---|---|---|---|
| Ea | 92.4±1.6 | 42.8±1.8 | 86.3±4.2 |
| Eb | 67.8±3.6 | 83.2±3.2 | 0.6±2.4 |
表2 酶对AFB1毒性位点相似结构化合物降解效果
Table 2 Degradation effects of enzymes on compounds with similar structures at the AFB1 toxicity site
| 酶 Enzyme | 香兰醛 Vanillin/% | 5-羟甲基糠醛 5-(Hydroxymethyl)furfural/% | 1-戊烯-3-酮 1-Penten-3-one/% |
|---|---|---|---|
| Ea | 92.4±1.6 | 42.8±1.8 | 86.3±4.2 |
| Eb | 67.8±3.6 | 83.2±3.2 | 0.6±2.4 |
| [1] |
Verheecke C, Liboz T, Mathieu F. Microbial degradation of aflatoxin B1: current status and future advances[J]. Int J Food Microbiol, 2016, 237: 1-9.
doi: S0168-1605(16)30386-5 pmid: 27541976 |
| [2] |
Guo YP, Zhao LH, Ma QG, et al. Novel strategies for degradation of aflatoxins in food and feed: a review[J]. Food Res Int, 2021, 140: 109878.
doi: 10.1016/j.foodres.2020.109878 URL |
| [3] |
Marin S, Ramos AJ, Cano-Sancho G, et al. Mycotoxins: occurrence, toxicology, and exposure assessment[J]. Food Chem Toxicol, 2013, 60: 218-237.
doi: 10.1016/j.fct.2013.07.047 pmid: 23907020 |
| [4] |
Adebo OA, Njobeh PB, Gbashi S, et al. Review on microbial degradation of aflatoxins[J]. Crit Rev Food Sci Nutr, 2017, 57(15): 3208-3217.
doi: 10.1080/10408398.2015.1106440 pmid: 26517507 |
| [5] |
Do JH, Choi DK. Aflatoxins: detection, toxicity, and biosynthesis[J]. Biotechnol Bioprocess Eng, 2007, 12(6): 585-593.
doi: 10.1007/BF02931073 URL |
| [6] |
赵萌, 高婧, 褚华硕, 等. 黄曲霉毒素B1的分子致毒机理及其微生物脱毒研究进展[J]. 食品科学, 2019, 40(11): 235-245.
doi: 10.7506/spkx1002-6630-20180502-001 |
| Zhao M, Gao J, Chu HS, et al. Recent progress in research on toxicity mechanism and microbial detoxification of aflatoxin B1[J]. Food Sci, 2019, 40(11): 235-245. | |
| [7] |
Cao H, Liu DL, Mo XM, et al. A fungal enzyme with the ability of aflatoxin B1 conversion: purification and ESI-MS/MS identification[J]. Microbiol Res, 2011, 166(6): 475-483.
doi: 10.1016/j.micres.2010.09.002 URL |
| [8] |
Wu YZ, Lu FP, Jiang HL, et al. The furofuran-ring selectivity, hydrogen peroxide-production and low Km value are the three elements for highly effective detoxification of aflatoxin oxidase[J]. Food Chem Toxicol, 2015, 76: 125-131.
doi: 10.1016/j.fct.2014.12.004 URL |
| [9] |
González Pereyra ML, Martínez MP, Cavaglieri LR. Presence of aiiA homologue genes encoding for N-Acyl homoserine lactone-degrading enzyme in aflatoxin B1-decontaminating Bacillus strains with potential use as feed additives[J]. Food Chem Toxicol, 2019, 124: 316-323.
doi: S0278-6915(18)30889-5 pmid: 30557671 |
| [10] |
Afsharmanesh H, Perez-Garcia A, Zeriouh H, et al. Aflatoxin degradation by Bacillus subtilis UTB1 is based on production of an oxidoreductase involved in bacilysin biosynthesis[J]. Food Control, 2018, 94: 48-55.
doi: 10.1016/j.foodcont.2018.03.002 URL |
| [11] | 唐璎, 邓展瑞, 黄佳, 等. 黄曲霉毒素B1降解菌株的鉴定及降解产物研究[J]. 食品与发酵工业, 2021, 47(7): 64-70. |
| Tang Y, Deng ZR, Huang J, et al. Screening and identification of an AFB1-dergrading strain and product analysis[J]. Food Ferment Ind, 2021, 47(7): 64-70. | |
| [12] | 宋茂鹏, 马现永, 邓盾, 等. 假单胞菌胞外酶降解黄曲霉毒素B1的酶学性质[J]. 微生物学通报, 2021, 48(1): 46-56. |
| Song MP, Ma XY, Deng D, et al. Characterization of an extracellular aflatoxin B1 degrading enzyme from a Pseudomonas strain[J]. Microbiol China, 2021, 48(1): 46-56. | |
| [13] | 田尔诺. 黄曲霉毒素B1降解酶及其降解机制研究[D]. 武汉: 湖北工业大学, 2018. |
| Tian EN. Study on the aflatoxin B1 degradation enzymes and degradation mechanism[D]. Wuhan: Hubei University of Technology, 2018. | |
| [14] |
Mwakinyali SE, Ming Z, Xie HL, et al. Investigation and characterization of Myroides odoratimimus strain 3J2MO aflatoxin B1 degradation[J]. J Agric Food Chem, 2019, 67(16): 4595-4602.
doi: 10.1021/acs.jafc.8b06810 URL |
| [15] | 唐璎, 孟宪刚, 邓展瑞, 等. 西北酸菜中吸附黄曲霉毒素B1乳酸菌株的筛选鉴定及稳定性研究[J]. 食品与发酵工业, 2020, 46(15): 60-65. |
| Tang Y, Meng XG, Deng ZR, et al. Screening and stability of an aflatoxin B1-adsorbing lactic acid bacterium isolated from pickled vegetable in northwest China[J]. Food Ferment Ind, 2020, 46(15): 60-65. | |
| [16] |
Glazunova OA, Polyakov KM, Moiseenko KV, et al. Structure-function study of two new middle-redox potential laccases from basidiomycetes Antrodiella faginea and Steccherinum murashkinskyi[J]. Int J Biol Macromol, 2018, 118(Pt A): 406-418.
doi: S0141-8130(18)30665-2 pmid: 29890251 |
| [17] |
唐璎, 黄佳, 邓展瑞, 等. 一株枯草芽孢杆菌降解黄曲霉毒素B1产物分析[J]. 生物技术通报, 2021, 37(12): 82-90.
doi: 10.13560/j.cnki.biotech.bull.1985.2021-0179 |
| Tang Y, Huang J, Deng ZR, et al. Product analysis of degrading aflatoxin B1 by a strain Bacillus subtilis[J]. Biotechnol Bull, 2021, 37(12): 82-90. | |
| [18] |
Mehrzad J, Malvandi AM, Alipour M, et al. Environmentally relevant level of aflatoxin B1 elicits toxic pro-inflammatory response in murine CNS-derived cells[J]. Toxicol Lett, 2017, 279: 96-106.
doi: S0378-4274(17)31157-8 pmid: 28789997 |
| [19] |
Alberts JF, Gelderblom WCA, Botha A, et al. Degradation of aflatoxin B(1)by fungal laccase enzymes[J]. Int J Food Microbiol, 2009, 135(1): 47-52.
doi: 10.1016/j.ijfoodmicro.2009.07.022 pmid: 19683355 |
| [20] |
Adebo OA, Njobeh PB, Sidu S, et al. Aflatoxin B1 degradation by liquid cultures and lysates of three bacterial strains[J]. Int J Food Microbiol, 2016, 233: 11-19.
doi: 10.1016/j.ijfoodmicro.2016.06.007 URL |
| [21] |
Guan S, Ji C, Zhou T, et al. Aflatoxin B(1)degradation by Stenotrophomonas maltophilia and other microbes selected using coumarin medium[J]. Int J Mol Sci, 2008, 9(8): 1489-1503.
doi: 10.3390/ijms9081489 pmid: 19325817 |
| [22] |
Chen GJ, Fang QA, Liao ZL, et al. Detoxification of aflatoxin B1 by a potential probiotic Bacillus amyloliquefaciens WF2020[J]. Front Microbiol, 2022, 13: 891091.
doi: 10.3389/fmicb.2022.891091 URL |
| [23] | Iram W, Anjum T, Iqbal M, et al. Structural analysis and biological toxicity of aflatoxins B1 and B2 degradation products following detoxification by Ocimum basilicum and Cassia fistula aqueous extracts[J]. Front Microbiol, 2016, 7: 1105. |
| [24] |
刘英丽, 毛慧佳, 杨梓妍, 等. 重组漆酶降解黄曲霉毒素B1分子对接分析及产物结构解析[J]. 食品科学, 2019, 40(24): 119-127.
doi: 10.7506/spkx1002-6630-20190623-260 |
|
Liu YL, Mao HJ, Yang ZY, et al. Molecular docking analysis of the interaction between recombinant laccase and aflatoxin B1 and structural elucidation of degradation products of aflatoxin B1[J]. Food Sci, 2019, 40(24): 119-127.
doi: 10.1111/jfds.1975.40.issue-1 URL |
|
| [25] |
Samuel MS, Sivaramakrishna A, Mehta A. Degradation and detoxification of aflatoxin B1 by Pseudomonas putida[J]. Int Biodeterior Biodegrad, 2014, 86: 202-209.
doi: 10.1016/j.ibiod.2013.08.026 URL |
| [26] |
Li JL, Huang J, Jin Y, et al. Mechanism and kinetics of degrading aflatoxin B1 by salt tolerant Candida versatilis CGMCC 3790[J]. J Hazard Mater, 2018, 359: 382-387.
doi: 10.1016/j.jhazmat.2018.05.053 URL |
| [27] | 阴佳璐. 浑浊红球菌PD630对黄曲霉毒素B1生物降解的研究[D]. 广州: 华南理工大学, 2020. |
| Yin JL. Study on biodegradation of aflatoxin B1 by Rhodococcus opacus PD630[D]. Guangzhou: South China University of Technology, 2020. | |
| [28] |
Christensen NJ, Kepp KP. Setting the stage for electron transfer: molecular basis of ABTS-binding to four laccases from Trametes versicolor at variable pH and protein oxidation state[J]. J Mol Catal B Enzym, 2014, 100: 68-77.
doi: 10.1016/j.molcatb.2013.11.017 URL |
| [29] |
刘玉灿, 段晋明, 李伟. 溶液pH值对二嗪磷紫外光降解产物及降解途径的影响[J]. 化学学报, 2015, 73(11): 1196-1202.
doi: 10.6023/A15070520 |
| Liu YC, Duan JM, Li W. Influence of solution pH on photolysis intermediates and degradation pathway of diazinon during UV irradiation treatment[J]. Acta Chimica Sin, 2015, 73(11): 1196-1202. | |
| [30] |
Fountain JC, Bajaj P, Nayak SN, et al. Responses of Aspergillus fla-vus to oxidative stress are related to fungal development regulator, antioxidant enzyme, and secondary metabolite biosynthetic gene expression[J]. Front Microbiol, 2016, 7: 2048.
doi: 10.3389/fmicb.2016.02048 pmid: 28066369 |
| [31] |
Casquete R, Benito MJ, Aranda E, et al. Gene expression of Aspergillus flavus strains on a cheese model system to control aflatoxin production[J]. J Dairy Sci, 2019, 102(9): 7765-7772.
doi: S0022-0302(19)30604-6 pmid: 31301828 |
| [32] |
Bai WZ, Feng TJ, Lan FX, et al. Study on the bio-function of lipA gene in Aspergillus flavus[J]. Genes Genomics, 2019, 41(1): 107-111.
doi: 10.1007/s13258-018-0744-7 |
| [33] | 丁炜. 黄曲霉毒素解毒酶基因的克隆与表达研究[D]. 呼和浩特: 内蒙古农业大学, 2010. |
| Ding W. The cloning and expression of gene encoding aflatoxin-detoxifizyme[D]. Hohhot: Inner Mongolia Agricultural University, 2010. |
| [1] | 赵志祥, 王殿东, 周亚林, 王培, 严婉荣, 严蓓, 罗路云, 张卓. 枯草芽孢杆菌Ya-1对辣椒枯萎病的防治及其对根际真菌群落的影响[J]. 生物技术通报, 2023, 39(9): 213-224. |
| [2] | 祖雪, 周瑚, 朱华珺, 任佐华, 刘二明. 枯草芽孢杆菌K-268的分离鉴定及对水稻稻瘟病的防病效果[J]. 生物技术通报, 2022, 38(6): 136-146. |
| [3] | 马艳琴, 邱益彬, 李莎, 徐虹. 透明质酸的生物合成及其代谢工程的研究进展[J]. 生物技术通报, 2022, 38(2): 252-262. |
| [4] | 张倩, 徐春燕, 张铎, 王亚会, 梁新盈, 李慧. 黄褐土玉米秸秆腐解菌株筛选及其促腐能力研究[J]. 生物技术通报, 2022, 38(12): 233-243. |
| [5] | 苗华彪, 曹艳, 杨梦瀚, 黄遵锡. 基于信号肽策略提高外源蛋白在枯草芽孢杆菌中的表达[J]. 生物技术通报, 2021, 37(6): 259-271. |
| [6] | 唐璎, 黄佳, 邓展瑞, 杨晓楠. 一株枯草芽孢杆菌降解黄曲霉毒素B1产物分析[J]. 生物技术通报, 2021, 37(12): 82-90. |
| [7] | 张维娇, 金学荣, 徐雅晴, 李江华, 堵国成, 康振. 枯草芽孢杆菌表达与调控工具相关研究进展[J]. 生物技术通报, 2020, 36(4): 26-33. |
| [8] | 付首颖, 夏苗苗, 张祎凝, 刘川, 涂然, 张大伟. 核黄素工业菌株高通量筛选方法的建立和应用[J]. 生物技术通报, 2020, 36(4): 47-53. |
| [9] | 赵晓霞, 牛世全, 文娜, 苏锋锋. 黄芪根腐病生防芽孢杆菌的筛选鉴定与盆栽防效试验[J]. 生物技术通报, 2019, 35(9): 107-111. |
| [10] | 邱锦, 黄火清, 姚斌, 罗会颖. 解淀粉芽孢杆菌淀粉酶催化活力改良及其在枯草芽孢杆菌中的高效表达[J]. 生物技术通报, 2019, 35(9): 134-143. |
| [11] | 凌翓, 纪明华, 段海燕, 史吉平, 孙俊松. 木糖酸氧化途径在枯草芽孢杆菌的构建及转化乙醇酸的研究[J]. 生物技术通报, 2019, 35(6): 76-82. |
| [12] | 戚家明, 杨娜, 孙杉杉, 明艳超, 郭亮, 张东旭, 徐志文. 一株具有噬菌体抗性的芽孢杆菌BS-2的鉴定及葡萄糖流加工艺优化[J]. 生物技术通报, 2019, 35(3): 210-216. |
| [13] | 戚家明, 孙杉杉, 张东旭, 徐志文, 徐延平. 芽孢杆菌BS-6基于全基因组数据的分类鉴定及拮抗 能力分析[J]. 生物技术通报, 2019, 35(10): 111-118. |
| [14] | 杨何宝 ,胡美荣 ,郑翔 ,牟庆璇 ,高沛汝. 不同信号肽及分子伴侣对中性蛋白酶在枯草芽孢杆菌中分泌表达的影响[J]. 生物技术通报, 2018, 34(6): 134-140. |
| [15] | 程新,李昆太,黄林. 一株枯草芽孢杆菌的生长特性及抑藻效果研究[J]. 生物技术通报, 2017, 33(7): 120-125. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
