








生物技术通报 ›› 2023, Vol. 39 ›› Issue (7): 185-194.doi: 10.13560/j.cnki.biotech.bull.1985.2022-1510
徐建霞1( ), 丁延庆1, 冯周1,2, 曹宁1, 程斌1, 高旭1, 邹桂花3, 张立异1(
), 丁延庆1, 冯周1,2, 曹宁1, 程斌1, 高旭1, 邹桂花3, 张立异1( )
)
收稿日期:2022-12-10
出版日期:2023-07-26
发布日期:2023-08-17
通讯作者:
张立异,女,博士,研究员,研究方向:作物分子遗传育种;E-mail: lyzhang1997@hotmail.com作者简介:徐建霞,男,硕士,助理研究员,研究方向:作物分子遗传育种;E-mail: 529438648 @qq.com
基金资助:
XU Jian-xia1( ), DING Yan-qing1, FENG Zhou1,2, CAO Ning1, CHENG Bin1, GAO Xu1, ZOU Gui-hua3, ZHANG Li-yi1(
), DING Yan-qing1, FENG Zhou1,2, CAO Ning1, CHENG Bin1, GAO Xu1, ZOU Gui-hua3, ZHANG Li-yi1( )
)
Received:2022-12-10
Published:2023-07-26
Online:2023-08-17
摘要:
株高及相关性状是影响高粱株型和产量的关键因素,定位影响这些性状的主效QTL,为高粱的分子遗传改良提供依据。以美国高粱品种BTx623和贵州酒用高粱品种红缨子杂交构建的包含205个家系的重组自交系(RIL)群体为材料,在5个环境条件下调查株高和节间数。利用Super-GBS技术进行基因分型,建立SNP标记遗传图谱,采用完备区间作图法(ICIM)开展QTL定位。结果表明,在第1、3、4、8、9染色体上一共检测到18个QTL,其中,与株高和节间数相关的QTL分别为7和11个。有10个主效QTL在多个环境中或2个性状中被重复检测到,其中qPH9.1与已知的株高基因Dw1一致,而qPH1.2、qPH3.2、qIN3.2以及qIN8.1所在区间内包含了4个与影响水稻株高或节间伸长的基因同源。
徐建霞, 丁延庆, 冯周, 曹宁, 程斌, 高旭, 邹桂花, 张立异. 基于Super-GBS的高粱株高和节间数QTL定位[J]. 生物技术通报, 2023, 39(7): 185-194.
XU Jian-xia, DING Yan-qing, FENG Zhou, CAO Ning, CHENG Bin, GAO Xu, ZOU Gui-hua, ZHANG Li-yi. QTL Mapping of Sorghum Plant Height and Internode Numbers Based on Super-GBS Technique[J]. Biotechnology Bulletin, 2023, 39(7): 185-194.
| 性状 Trait | 环境 Environment | 亲本Parents | RIL群体RIL population | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BTx623 | 红缨子 Hongyingzi | 最小值 Minimum | 最大值 Maximum | 平均值 Mean | 标准差 SD | 变异系数 CV/% | 偏度 Skewness | 峰度 Kurtosis | ||||
| 株高 PH/cm | 2020GY | 120.33 | 266.33 | 101.00 | 360.67 | 207.37 | 56.94 | 27.46 | 0.176 | -0.769 | ||
| 2020AS | 132.67 | 284.00 | 106.67 | 320.33 | 197.99 | 47.66 | 24.07 | 0.192 | -0.773 | |||
| 2020LD | 122.00 | 196.33 | 81.67 | 254.00 | 178.02 | 31.65 | 17.78 | -0.213 | -0.453 | |||
| 2021GY | 124.33 | 215.00 | 105.00 | 316.67 | 201.16 | 43.32 | 21.53 | 0.070 | -0.513 | |||
| 2021AS | 147.00 | 238.67 | 107.33 | 321.67 | 196.15 | 42.99 | 21.92 | 0.188 | -0.671 | |||
| 节间数 IN | 2020GY | 8.67 | 10.67 | 5.67 | 13.33 | 9.73 | 1.19 | 12.22 | 0.144 | 0.404 | ||
| 2020AS | 7.67 | 10.33 | 6.00 | 12.67 | 8.75 | 1.27 | 14.52 | 0.509 | -0.248 | |||
| 2020LD | 7.00 | 9.33 | 4.67 | 11.00 | 7.48 | 0.98 | 13.07 | 0.402 | 1.609 | |||
| 2021GY | 7.00 | 8.67 | 5.33 | 11.00 | 8.19 | 0.98 | 12.02 | 0.016 | 0.673 | |||
| 2021AS | 7.00 | 9.67 | 5.33 | 12.67 | 8.49 | 1.41 | 16.64 | 0.377 | -0.087 | |||
表1 RIL群体中株高和节间数在5个环境下的表型统计
Table 1 Phenotypic statistics of plant height and internode numbers in the RIL population in five environments
| 性状 Trait | 环境 Environment | 亲本Parents | RIL群体RIL population | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BTx623 | 红缨子 Hongyingzi | 最小值 Minimum | 最大值 Maximum | 平均值 Mean | 标准差 SD | 变异系数 CV/% | 偏度 Skewness | 峰度 Kurtosis | ||||
| 株高 PH/cm | 2020GY | 120.33 | 266.33 | 101.00 | 360.67 | 207.37 | 56.94 | 27.46 | 0.176 | -0.769 | ||
| 2020AS | 132.67 | 284.00 | 106.67 | 320.33 | 197.99 | 47.66 | 24.07 | 0.192 | -0.773 | |||
| 2020LD | 122.00 | 196.33 | 81.67 | 254.00 | 178.02 | 31.65 | 17.78 | -0.213 | -0.453 | |||
| 2021GY | 124.33 | 215.00 | 105.00 | 316.67 | 201.16 | 43.32 | 21.53 | 0.070 | -0.513 | |||
| 2021AS | 147.00 | 238.67 | 107.33 | 321.67 | 196.15 | 42.99 | 21.92 | 0.188 | -0.671 | |||
| 节间数 IN | 2020GY | 8.67 | 10.67 | 5.67 | 13.33 | 9.73 | 1.19 | 12.22 | 0.144 | 0.404 | ||
| 2020AS | 7.67 | 10.33 | 6.00 | 12.67 | 8.75 | 1.27 | 14.52 | 0.509 | -0.248 | |||
| 2020LD | 7.00 | 9.33 | 4.67 | 11.00 | 7.48 | 0.98 | 13.07 | 0.402 | 1.609 | |||
| 2021GY | 7.00 | 8.67 | 5.33 | 11.00 | 8.19 | 0.98 | 12.02 | 0.016 | 0.673 | |||
| 2021AS | 7.00 | 9.67 | 5.33 | 12.67 | 8.49 | 1.41 | 16.64 | 0.377 | -0.087 | |||
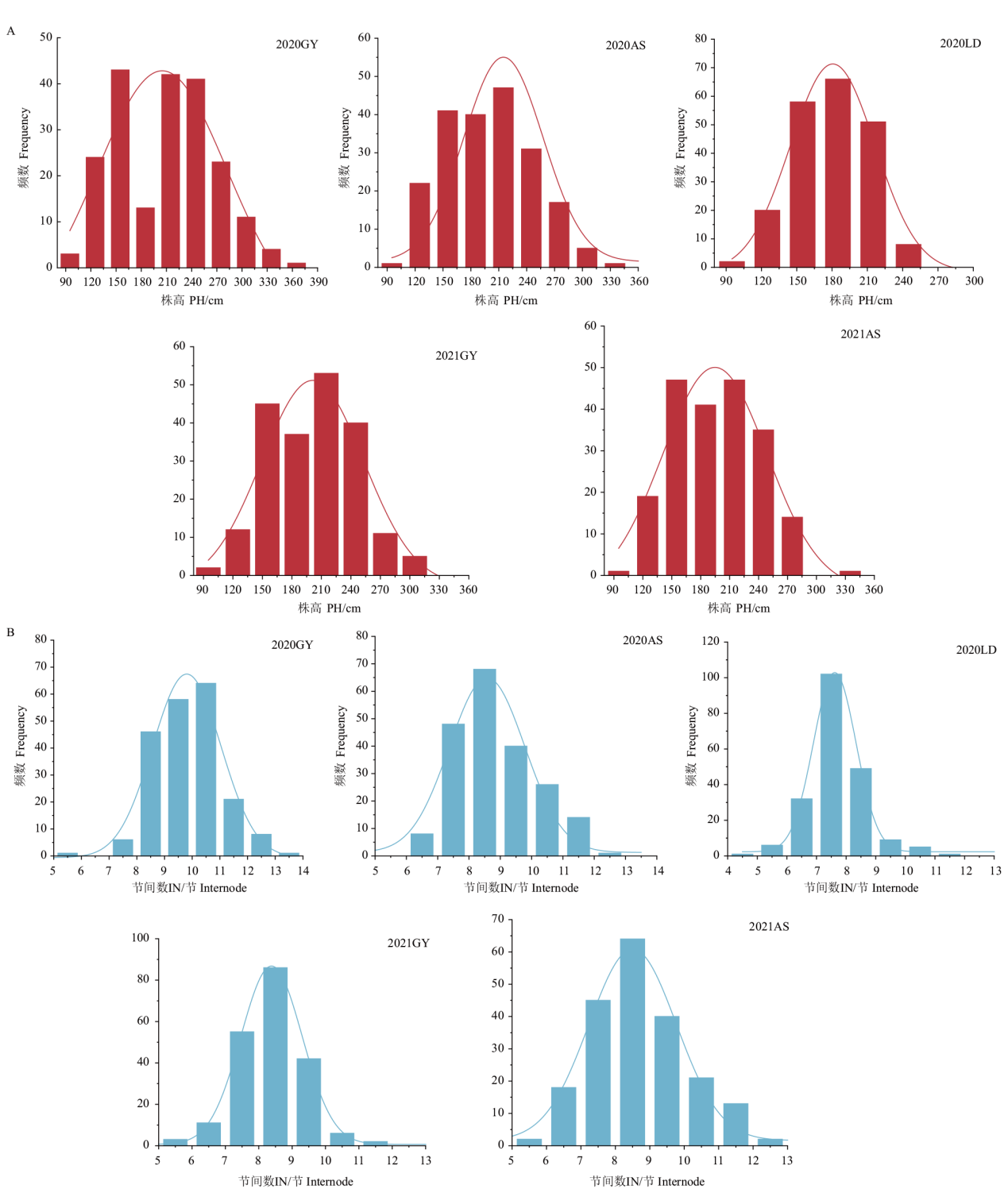
图1 RIL群体在5个环境(2020贵阳、安顺和乐东,以及2021年贵阳和安顺)中株高(A)和节间数(B)表型数据频数分布图
Fig. 1 Frequency distribution of plant heights(A)and internode numbers(B) in RIL population in five environments of Guiyang(2020GY),Anshun(2020AS)and Ledong(2020LD)in 2020, as well as Guiyang(2021GY)and Anshun(2021AS)in 2021
| 性状Trait | 2020GY-PH | 2020AS-PH | 2020SY-PH | 2021GY-PH | 2021AS-PH | 2020GY-IN | 2020AS-IN | 2020SY-IN | 2021GY-IN | 2021AS-IN |
|---|---|---|---|---|---|---|---|---|---|---|
| 2020GY-PH | 1.000 | |||||||||
| 2020AS-PH | 0.832** | 1.000 | ||||||||
| 2020SY-PH | 0.884** | 0.783** | 1.000 | |||||||
| 2021GY-PH | 0.860** | 0.826** | 0.856** | 1.000 | ||||||
| 2021AS-PH | 0.844** | 0.760** | 0.814** | 0.861** | 1.000 | |||||
| 2020GY-IN | 0.474** | 0.457** | 0.387** | 0.418** | 0.381** | 1.000 | ||||
| 2020AS-IN | 0.399** | 0.584** | 0.317** | 0.383** | 0.380** | 0.651** | 1.000 | |||
| 2020SY-IN | 0.431** | 0.422** | 0.491** | 0.428** | 0.367** | 0.671** | 0.500** | 1.000 | ||
| 2021GY-IN | 0.380** | 0.472** | 0.302** | 0.472** | 0.390** | 0.684** | 0.665** | 0.575** | 1.000 | |
| 2021AS-IN | 0.475** | 0.512** | 0.432** | 0.515** | 0.618** | 0.567** | 0.633** | 0.508** | 0.599** | 1.000 |
表2 RIL群体中株高和节间数间的相关性分析
Table 2 Correlation analysis of plant heighst and internode numbers in the RIL population
| 性状Trait | 2020GY-PH | 2020AS-PH | 2020SY-PH | 2021GY-PH | 2021AS-PH | 2020GY-IN | 2020AS-IN | 2020SY-IN | 2021GY-IN | 2021AS-IN |
|---|---|---|---|---|---|---|---|---|---|---|
| 2020GY-PH | 1.000 | |||||||||
| 2020AS-PH | 0.832** | 1.000 | ||||||||
| 2020SY-PH | 0.884** | 0.783** | 1.000 | |||||||
| 2021GY-PH | 0.860** | 0.826** | 0.856** | 1.000 | ||||||
| 2021AS-PH | 0.844** | 0.760** | 0.814** | 0.861** | 1.000 | |||||
| 2020GY-IN | 0.474** | 0.457** | 0.387** | 0.418** | 0.381** | 1.000 | ||||
| 2020AS-IN | 0.399** | 0.584** | 0.317** | 0.383** | 0.380** | 0.651** | 1.000 | |||
| 2020SY-IN | 0.431** | 0.422** | 0.491** | 0.428** | 0.367** | 0.671** | 0.500** | 1.000 | ||
| 2021GY-IN | 0.380** | 0.472** | 0.302** | 0.472** | 0.390** | 0.684** | 0.665** | 0.575** | 1.000 | |
| 2021AS-IN | 0.475** | 0.512** | 0.432** | 0.515** | 0.618** | 0.567** | 0.633** | 0.508** | 0.599** | 1.000 |
| 性状 Trait | 均方 Mean square | F值F value | 广义遗传率 h2B/% | |||||
|---|---|---|---|---|---|---|---|---|
| 基因型Genotype | 环境Environment | 基因型iro G×E | 基因型Genotype | 环境Environment | 基因型iro G×E | |||
| 株高PH | 26 001.18 | 75 159.27 | 1 238.43 | 224.52** | 649.01** | 10.69** | 95.23 | |
| 节间数IN | 14.05 | 419.71 | 1.70 | 43.24** | 1 291.97** | 5.23** | 85.78 | |
表3 株高和节间数间方差分析及广义遗传率
Table 3 Variance analysis and broad heritability of plant height and internode numbers
| 性状 Trait | 均方 Mean square | F值F value | 广义遗传率 h2B/% | |||||
|---|---|---|---|---|---|---|---|---|
| 基因型Genotype | 环境Environment | 基因型iro G×E | 基因型Genotype | 环境Environment | 基因型iro G×E | |||
| 株高PH | 26 001.18 | 75 159.27 | 1 238.43 | 224.52** | 649.01** | 10.69** | 95.23 | |
| 节间数IN | 14.05 | 419.71 | 1.70 | 43.24** | 1 291.97** | 5.23** | 85.78 | |
| 性状 Trait | 染色体 Chr. | 位点 QTL | 环境 Env. | 位置 Pos./cM | 标记区间 Marker interval | 物理距离 Phy./Mb | 阈值 LOD | 贡献率 PVE/% | 加性效应 Add |
|---|---|---|---|---|---|---|---|---|---|
| 株高 PH | 1 | qPH1.1 | 2021AS | 19.50 | Sb01819-Sb01824 | 19.72-20.13 | 5.12 | 5.12 | -10.43 |
| qPH1.2 | 2021AS | 29.40 | Sb01925-Sb01957 | 25.81-27.00 | 10.14 | 15.19 | 15.19 | ||
| 2020GY | 30.90 | Sb01939-Sb01941 | 26.58-26.58 | 2.68 | 2.82 | 9.10 | |||
| 3 | qPH3.1 | 2020GY | 63.50 | Sb035554-Sb035557 | 56.08-56.14 | 8.58 | 9.13 | -13.99 | |
| 2020AS | 63.50 | Sb035554-Sb035557 | 56.08-56.14 | 4.39 | 4.39 | -10.23 | |||
| qPH3.2 | 2021GY | 65.30 | Sb035584-Sb035595 | 56.41-56.90 | 9.19 | 10.72 | -15.19 | ||
| 2021AS | 65.30 | Sb035584-Sb035595 | 56.41-56.90 | 4.15 | 4.46 | -11.39 | |||
| 4 | qPH4.1 | 2020AS | 0.50 | Sb046572-Sb046576 | 1.48-1.53 | 5.83 | 6.66 | -12.04 | |
| 2021AS | 0.50 | Sb046572-Sb046576 | 1.48-1.53 | 4.00 | 4.01 | -9.30 | |||
| 2021GY | 0.50 | Sb046572-Sb046576 | 1.48-1.53 | 3.70 | 3.55 | -9.25 | |||
| qPH4.2 | 2020GY | 3.50 | Sb046608-Sb046614 | 2.01-2.27 | 5.15 | 5.58 | -12.81 | ||
| 9 | qPH9.1 | 2020AS | 85.90 | Sb0915270-Sb0915408 | 54.55-58.47 | 6.98 | 11.36 | -15.74 | |
| 2021AS | 85.50 | Sb0915270-Sb0915408 | 54.55-58.47 | 4.35 | 5.98 | -11.39 | |||
| 2020GY | 86.20 | Sb0915270-Sb0915408 | 54.55-58.47 | 7.07 | 11.30 | -18.27 | |||
| 2020GY | 93.60 | Sb0915373-Sb0915384 | 57.67-57.85 | 24.84 | 34.34 | -31.69 | |||
| 2020AS | 93.60 | Sb0915373-Sb0915384 | 57.67-57.85 | 20.75 | 27.82 | -24.51 | |||
| 2021AS | 93.70 | Sb0915373-Sb0915384 | 57.67-57.85 | 15.95 | 18.25 | -19.78 | |||
| 节间数 IN | 1 | qIN1.1 | 2020LD | 53.60 | Sb011709-Sb011710 | 62.62-62.71 | 3.03 | 4.19 | 0.22 |
| 3 | qIN3.1 | 2020AS | 59.30 | Sb035466-Sb035520 | 53.57-55.06 | 19.22 | 23.92 | -0.72 | |
| 2021AS | 60.00 | Sb035466-Sb035520 | 53.57-55.06 | 10.89 | 18.64 | -0.62 | |||
| qIN3.2 | 2020GY | 65.10 | Sb035598-Sb035611 | 56.91-57.32 | 5.29 | 7.98 | -0.33 | ||
| 2021GY | 65.40 | Sb035560-Sb035597 | 56.23-56.91 | 6.84 | 11.90 | -1.33 | |||
| 4 | qIN4.1 | 2020AS | 0.50 | Sb046572-Sb046576 | 1.48-1.53 | 6.05 | 6.37 | -0.37 | |
| qIN4.2 | 2020LD | 60.90 | Sb047891-Sb047937 | 55.91-57.11 | 3.24 | 4.99 | -0.24 | ||
| qIN4.3 | 2020AS | 70.20 | Sb048094-Sb048097 | 60.95-61.09 | 2.55 | 2.56 | -0.23 | ||
| 8 | qIN8.1 | 2020GY | 76.50 | Sb0813812-Sb0813988 | 58.25-60.76 | 4.30 | 8.77 | 0.35 | |
| 2020LD | 76.20 | Sb0813812-Sb0813988 | 58.25-60.76 | 7.90 | 14.84 | 0.41 | |||
| qIN8.2 | 2020AS | 85.30 | Sb0814059-Sb0814062 | 62.02-62.05 | 2.96 | 2.96 | 0.25 | ||
| 9 | qIN9.1 | 2021GY | 77.50 | Sb0915233-Sb0915254 | 52.93-53.94 | 3.84 | 6.99 | -0.39 | |
| 2021AS | 79.60 | Sb0915254-Sb0915270 | 53.94-54.55 | 4.20 | 6.73 | -0.37 | |||
| 2020GY | 81.50 | Sb0915254-Sb0915270 | 53.94-54.55 | 4.45 | 6.60 | -0.31 | |||
| qIN9.2 | 2020LD | 93.50 | Sb0915373-Sb0915384 | 57.67-57.85 | 9.15 | 13.47 | -0.39 | ||
| qIN9.3 | 2020GY | 98.30 | Sb0915437-Sb0915441 | 58.96-58.99 | 6.75 | 10.05 | -0.37 | ||
| 2020AS | 97.70 | Sb0915434-Sb0915435 | 58.82-58.88 | 7.92 | 8.57 | -0.43 | |||
| 2021AS | 97.90 | Sb0915435-Sb0915437 | 58.88-58.96 | 4.62 | 7.18 | -0.38 |
表4 株高和节间数相关QTL定位结果
Table 4 QTL mapping results of plant height and internode numbers
| 性状 Trait | 染色体 Chr. | 位点 QTL | 环境 Env. | 位置 Pos./cM | 标记区间 Marker interval | 物理距离 Phy./Mb | 阈值 LOD | 贡献率 PVE/% | 加性效应 Add |
|---|---|---|---|---|---|---|---|---|---|
| 株高 PH | 1 | qPH1.1 | 2021AS | 19.50 | Sb01819-Sb01824 | 19.72-20.13 | 5.12 | 5.12 | -10.43 |
| qPH1.2 | 2021AS | 29.40 | Sb01925-Sb01957 | 25.81-27.00 | 10.14 | 15.19 | 15.19 | ||
| 2020GY | 30.90 | Sb01939-Sb01941 | 26.58-26.58 | 2.68 | 2.82 | 9.10 | |||
| 3 | qPH3.1 | 2020GY | 63.50 | Sb035554-Sb035557 | 56.08-56.14 | 8.58 | 9.13 | -13.99 | |
| 2020AS | 63.50 | Sb035554-Sb035557 | 56.08-56.14 | 4.39 | 4.39 | -10.23 | |||
| qPH3.2 | 2021GY | 65.30 | Sb035584-Sb035595 | 56.41-56.90 | 9.19 | 10.72 | -15.19 | ||
| 2021AS | 65.30 | Sb035584-Sb035595 | 56.41-56.90 | 4.15 | 4.46 | -11.39 | |||
| 4 | qPH4.1 | 2020AS | 0.50 | Sb046572-Sb046576 | 1.48-1.53 | 5.83 | 6.66 | -12.04 | |
| 2021AS | 0.50 | Sb046572-Sb046576 | 1.48-1.53 | 4.00 | 4.01 | -9.30 | |||
| 2021GY | 0.50 | Sb046572-Sb046576 | 1.48-1.53 | 3.70 | 3.55 | -9.25 | |||
| qPH4.2 | 2020GY | 3.50 | Sb046608-Sb046614 | 2.01-2.27 | 5.15 | 5.58 | -12.81 | ||
| 9 | qPH9.1 | 2020AS | 85.90 | Sb0915270-Sb0915408 | 54.55-58.47 | 6.98 | 11.36 | -15.74 | |
| 2021AS | 85.50 | Sb0915270-Sb0915408 | 54.55-58.47 | 4.35 | 5.98 | -11.39 | |||
| 2020GY | 86.20 | Sb0915270-Sb0915408 | 54.55-58.47 | 7.07 | 11.30 | -18.27 | |||
| 2020GY | 93.60 | Sb0915373-Sb0915384 | 57.67-57.85 | 24.84 | 34.34 | -31.69 | |||
| 2020AS | 93.60 | Sb0915373-Sb0915384 | 57.67-57.85 | 20.75 | 27.82 | -24.51 | |||
| 2021AS | 93.70 | Sb0915373-Sb0915384 | 57.67-57.85 | 15.95 | 18.25 | -19.78 | |||
| 节间数 IN | 1 | qIN1.1 | 2020LD | 53.60 | Sb011709-Sb011710 | 62.62-62.71 | 3.03 | 4.19 | 0.22 |
| 3 | qIN3.1 | 2020AS | 59.30 | Sb035466-Sb035520 | 53.57-55.06 | 19.22 | 23.92 | -0.72 | |
| 2021AS | 60.00 | Sb035466-Sb035520 | 53.57-55.06 | 10.89 | 18.64 | -0.62 | |||
| qIN3.2 | 2020GY | 65.10 | Sb035598-Sb035611 | 56.91-57.32 | 5.29 | 7.98 | -0.33 | ||
| 2021GY | 65.40 | Sb035560-Sb035597 | 56.23-56.91 | 6.84 | 11.90 | -1.33 | |||
| 4 | qIN4.1 | 2020AS | 0.50 | Sb046572-Sb046576 | 1.48-1.53 | 6.05 | 6.37 | -0.37 | |
| qIN4.2 | 2020LD | 60.90 | Sb047891-Sb047937 | 55.91-57.11 | 3.24 | 4.99 | -0.24 | ||
| qIN4.3 | 2020AS | 70.20 | Sb048094-Sb048097 | 60.95-61.09 | 2.55 | 2.56 | -0.23 | ||
| 8 | qIN8.1 | 2020GY | 76.50 | Sb0813812-Sb0813988 | 58.25-60.76 | 4.30 | 8.77 | 0.35 | |
| 2020LD | 76.20 | Sb0813812-Sb0813988 | 58.25-60.76 | 7.90 | 14.84 | 0.41 | |||
| qIN8.2 | 2020AS | 85.30 | Sb0814059-Sb0814062 | 62.02-62.05 | 2.96 | 2.96 | 0.25 | ||
| 9 | qIN9.1 | 2021GY | 77.50 | Sb0915233-Sb0915254 | 52.93-53.94 | 3.84 | 6.99 | -0.39 | |
| 2021AS | 79.60 | Sb0915254-Sb0915270 | 53.94-54.55 | 4.20 | 6.73 | -0.37 | |||
| 2020GY | 81.50 | Sb0915254-Sb0915270 | 53.94-54.55 | 4.45 | 6.60 | -0.31 | |||
| qIN9.2 | 2020LD | 93.50 | Sb0915373-Sb0915384 | 57.67-57.85 | 9.15 | 13.47 | -0.39 | ||
| qIN9.3 | 2020GY | 98.30 | Sb0915437-Sb0915441 | 58.96-58.99 | 6.75 | 10.05 | -0.37 | ||
| 2020AS | 97.70 | Sb0915434-Sb0915435 | 58.82-58.88 | 7.92 | 8.57 | -0.43 | |||
| 2021AS | 97.90 | Sb0915435-Sb0915437 | 58.88-58.96 | 4.62 | 7.18 | -0.38 |

图2 高密度遗传图谱及重要QTL所在染色体示意图 1个环境检测到的QTL显示为黑色,2个及以上环境检测到的QTL显示为红色
Fig. 2 Schematic diagram of high-density genetic map and chromosomes with important QTL regions QTLs identified in one environment and in two or more environments are shown in black and red, respectively
| QTL | 候选基因 Candidate gene | 基因组位置 Genomic location | 同源基因 Orthologous gene | 蛋白注释 Proteins annotation | 相似性 Similarity/% |
|---|---|---|---|---|---|
| qPH1.2 | Sobic001G248600 | Chr01:26782638-26792923 | OsCPS1(LOC_Os02g17780) | 对映型柯巴基焦磷酸合酶 Ent-copalyl diphosphate synthase | 82.30 |
| qPH3.2/qIN3.2 | Sobic003G227000 | Chr03:56425156-56426899 | OsOFP2(LOC_Os01g43610) | 卵形家族蛋白 Ovate family protein | 68.20 |
| qPH3.2 | Sobic003G227300 | Chr03:56472263-56479136 | Dlf1(LOC_Os01g43650) | WRKY转录因子 WRKY transcription factor | 76.40 |
| qPH9.1/qIN9.2 | Sobic.009G230800/DW1 | Chr09:57093313-57095643 | OsGA2ox(LOC_Os05g48700) | 赤霉素2氧化酶 Gibberellin 2-oxidase gene | 87.00 |
| qIN8.1 | Sobic008G173300 | Chr08:60694532-60695140 | DEC1(LOC_Os12g42250) | 节间伸长抑制因子;C2H2锌指转录因子 Decelerator of internode elongation; Cys2His2(C2H2)zinc-finger transcription factor | 56.00 |
表5 重要QTL区间候选基因功能注释
Table 5 Functional annotation of candidate genes in important QTL intervals
| QTL | 候选基因 Candidate gene | 基因组位置 Genomic location | 同源基因 Orthologous gene | 蛋白注释 Proteins annotation | 相似性 Similarity/% |
|---|---|---|---|---|---|
| qPH1.2 | Sobic001G248600 | Chr01:26782638-26792923 | OsCPS1(LOC_Os02g17780) | 对映型柯巴基焦磷酸合酶 Ent-copalyl diphosphate synthase | 82.30 |
| qPH3.2/qIN3.2 | Sobic003G227000 | Chr03:56425156-56426899 | OsOFP2(LOC_Os01g43610) | 卵形家族蛋白 Ovate family protein | 68.20 |
| qPH3.2 | Sobic003G227300 | Chr03:56472263-56479136 | Dlf1(LOC_Os01g43650) | WRKY转录因子 WRKY transcription factor | 76.40 |
| qPH9.1/qIN9.2 | Sobic.009G230800/DW1 | Chr09:57093313-57095643 | OsGA2ox(LOC_Os05g48700) | 赤霉素2氧化酶 Gibberellin 2-oxidase gene | 87.00 |
| qIN8.1 | Sobic008G173300 | Chr08:60694532-60695140 | DEC1(LOC_Os12g42250) | 节间伸长抑制因子;C2H2锌指转录因子 Decelerator of internode elongation; Cys2His2(C2H2)zinc-finger transcription factor | 56.00 |
| [1] |
李顺国, 刘猛, 刘斐, 等. 中国高粱产业和种业发展现状与未来展望[J]. 中国农业科学, 2021, 54(3): 471-482.
doi: 10.3864/j.issn.0578-1752.2021.03.002 |
|
Li SG, Liu M, Liu F, et al. Current status and future prospective of sorghum production and seed industry in China[J]. Sci Agric Sin, 2021, 54(3): 471-482.
doi: 10.3864/j.issn.0578-1752.2021.03.002 |
|
| [2] | 仪治本, 梁小红, 赵威军, 等. 高粱基因组遗传图谱构建的研究进展[J]. 农业生物技术学报, 2006, 14(2): 279-285. |
| Yi ZB, Liang XH, Zhao WJ, et al. Advances in genetic mapping of sorghum genome[J]. J Agric Biotechnol, 2006, 14(2): 279-285. | |
| [3] | 高士杰, 刘晓辉, 李继洪. 高粱高产育种应重视株型和穗结构性状的改良[J]. 种子, 2007, 26(3): 83-84. |
| Gao SJ, Liu XH, Li JH. Paying much attention to the improvement of plant type and spike structure in high-yielding breeding of sorghum[J]. Seed, 2007, 26(3): 83-84. | |
| [4] | Baggett JP, Tillett RL, Cooper EA, et al. De novo identification and targeted sequencing of SSRs efficiently fingerprints sorghum bicolor sub-population identity[J]. PLoS One, 2021, 16(3): e0248213. |
| [5] |
Wang LM, Jiao SJ, Jiang YX, et al. Genetic diversity in parent lines of sweet sorghum based on agronomical traits and SSR markers[J]. Field Crops Res, 2013, 149: 11-19.
doi: 10.1016/j.fcr.2013.04.013 URL |
| [6] |
Reddy RN, Madhusudhana R, Mohan SM, et al. Mapping QTL for grain yield and other agronomic traits in post-rainy sorghum[Sor-ghum bicolor(L.) Moench[J]. Theor Appl Genet, 2013, 126(8): 1921-1939.
doi: 10.1007/s00122-013-2107-8 URL |
| [7] | 苏舒. 高粱形态学农艺性状的QTL定位研究[D]. 南京: 南京大学, 2012. |
| Su S. QTL mapping of agronomic traits of morphology in sorghum[D]. Nanjing: Nanjing University, 2012. | |
| [21] |
Ding YQ, Xu JX, Wang C, et al. QTL mapping of grain traits related to brewing in sorghum based on super-GBS technology[J]. J Nucl Agric Sci, 2023, 37(2): 241-250.
doi: 10.11869/j.issn.1000-8551.2023.02.0241 |
| [22] |
Meng L, Li HH, Zhang LY, et al. QTL IciMapping: integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations[J]. Crop J, 2015, 3(3): 269-283.
doi: 10.1016/j.cj.2015.01.001 |
| [23] |
丁延庆, 周棱波, 汪灿, 等. 酱香型酒用糯高粱研究进展[J]. 生物技术通报, 2019, 35(5): 28-34.
doi: 10.13560/j.cnki.biotech.bull.1985.2019-0080 URL |
| Ding YQ, Zhou LB, Wang C, et al. Research advance in glutinous sorghum for making sauce-flavor liquor in China[J]. Biotechnol Bull, 2019, 35(5): 28-34. | |
| [24] | 中商情报网. 2020年贵州省酒用高粱市场供需形势及产量预测分析[EB/OL].(2020-7-22)[2022-12-03]. https://www.163.com/dy/article/FI5ICNRR051481OF.html. |
| China Business Information Network. Market supply and demand situation and yield forecast analysis of wine sorghum in Guizhou province in 2020[EB/OL].(2020-7-22)[2022-12-03]. https://www.163.com/dy/article/FI5ICNRR051481OF.html. | |
| [25] | 丁延庆, 曹宁, 周棱波, 等. 酒用糯高粱EMS突变体库构建及突变体筛选[J]. 南方农业学报, 2020, 51(12): 2884-2891. |
| Ding YQ, Cao N, Zhou LB, et al. Construction of EMS-induced mutant library and mutant screening in liquor-making waxy sorghum[J]. J South Agric, 2020, 51(12): 2884-2891. | |
| [26] |
郑海洋, 侯立龙, 曹富斌, 等. 大豆株高QTL定位及候选基因挖掘[J/OL]. 中国油料作物学报, 2022. DOI: 10.19802/j.issn.1007-9084.2022065.
doi: 10.19802/j.issn.1007-9084.2022065 |
|
Zheng HY, Hou LL, Cao FB, et al. Soybean plant height QTL mapping and candidate gene mining[J/OL]. Chin J Oil Crop Sci, 2022. DOI: 10.19802/j.iSSN.1007-9084.2022065.
doi: 10.19802/j.iSSN.1007-9084.2022065 |
|
| [27] | Zhao J, Mantilla Perez MB, Hu JY, et al. Genome-wide association study for nine plant architecture traits in Sorghum[J]. Plant Genome, 2016, 9(2): plantgenome2015.06.0044. |
| [8] | 王丽华, 陆叶飞, 陆以静, 等. “高粱不育系Tx623A×苏丹草Sa”RIL群体农艺性状的QTL定位[J]. 山西农业科学, 2021, 49(12): 1491-1496. |
| Wang LH, Lu YF, Lu YJ, et al. QTL mapping of the agronomic traits using an RIL populations of “Tx623A × sa”[J]. J Shanxi Agric Sci, 2021, 49(12): 1491-1496. | |
| [9] |
Zou GH, Zhai GW, Feng Q, et al. Identification of QTLs for eight agronomically important traits using an ultra-high-density map based on SNPs generated from high-throughput sequencing in sorghum under contrasting photoperiods[J]. J Exp Bot, 2012, 63(15): 5451-5462.
doi: 10.1093/jxb/ers205 pmid: 22859680 |
| [10] |
Kajiya-Kanegae H, Takanashi H, Fujimoto M, et al. RAD-seq-based high-density linkage map construction and QTL mapping of biomass-related traits in sorghum using the Japanese Landrace takakibi NOG[J]. Plant Cell Physiol, 2020, 61(7): 1262-1272.
doi: 10.1093/pcp/pcaa056 pmid: 32353144 |
| [11] |
Takanashi H, Shichijo M, Sakamoto L, et al. Genetic dissection of QTLs associated with spikelet-related traits and grain size in sorghum[J]. Sci Rep, 2021, 11(1): 9398.
doi: 10.1038/s41598-021-88917-x pmid: 33931706 |
| [12] |
Brown PJ, Rooney WL, Franks C, et al. Efficient mapping of plant height quantitative trait loci in a sorghum association population with introgressed dwarfing genes[J]. Genetics, 2008, 180(1): 629-637.
doi: 10.1534/genetics.108.092239 pmid: 18757942 |
| [13] |
Morris GP, Ramu P, Deshpande SP, et al. Population genomic and genome-wide association studies of agroclimatic traits in sorghum[J]. Proc Natl Acad Sci USA, 2013, 110(2): 453-458.
doi: 10.1073/pnas.1215985110 pmid: 23267105 |
| [14] |
Hirano K, Kawamura M, Araki-Nakamura S, et al. Sorghum DW1 positively regulates brassinosteroid signaling by inhibiting the nuclear localization of BRASSINOSTEROID INSENSITIVE 2[J]. Sci Rep, 2017, 7(1): 126.
doi: 10.1038/s41598-017-00096-w pmid: 28273925 |
| [15] |
Hilley JL, Weers BD, Truong SK, et al. Sorghum Dw2 encodes a protein kinase regulator of stem internode length[J]. Sci Rep, 2017, 7(1): 4616.
doi: 10.1038/s41598-017-04609-5 pmid: 28676627 |
| [16] |
Li X, Li XR, Fridman E, et al. Dissecting repulsion linkage in the dwarfing gene Dw3 region for sorghum plant height provides insights into heterosis[J]. Proc Natl Acad Sci USA, 2015, 112(38): 11823-11828.
doi: 10.1073/pnas.1509229112 pmid: 26351684 |
| [28] |
Hart GE, Schertz KF, Peng Y, et al. Genetic mapping of Sorghum bicolor(L.) Moench QTLs that control variation in tillering and other morphological characters[J]. Theor Appl Genet, 2001, 103(8): 1232-1242.
doi: 10.1007/s001220100582 URL |
| [29] | Otomo K, Kenmoku H, Oikawa H, et al. Biological functions of ent- and syn-copalyl diphosphate synthases in rice: key enzymes for the branch point of gibberellin and phytoalexin biosynthesis[J]. PlantJ, 2004, 39(6): 886-893. |
| [30] |
Bensen RJ, Johal GS, Crane VC, et al. Cloning and characterization of the maize An1 gene[J]. Plant Cell, 1995, 7(1): 75-84.
doi: 10.1105/tpc.7.1.75 pmid: 7696880 |
| [31] |
Shiringani AL, Frisch M, Friedt W. Genetic mapping of QTLs for sugar-related traits in a RIL population of Sorghum bicolor L. Moench[J]. Theor Appl Genet, 2010, 121(2): 323-336.
doi: 10.1007/s00122-010-1312-y pmid: 20229249 |
| [32] |
Liu HH, Liu HQ, Zhou LN, et al. Genetic architecture of domestication- and improvement-related traits using a population derived from Sorghum virgatum and Sorghum bicolor[J]. Plant Sci, 2019, 283: 135-146.
doi: 10.1016/j.plantsci.2019.02.013 URL |
| [33] |
Felderhoff TJ, Murray SC, Klein PE, et al. QTLs for energy-related traits in a sweet × grain sorghum[Sorghum bicolor(L.) Moench]mapping population[J]. Crop Sci, 2012, 52(5): 2040-2049.
doi: 10.2135/cropsci2011.11.0618 URL |
| [34] |
Nagai K, Mori Y, Ishikawa S, et al. Antagonistic regulation of the gibberellic acid response during stem growth in rice[J]. Nature, 2020, 584(7819): 109-114.
doi: 10.1038/s41586-020-2501-8 |
| [35] |
Schmitz AJ, Begcy K, Sarath G, et al. Rice Ovate Family Protein 2(OFP2)alters hormonal homeostasis and vasculature development[J]. Plant Sci, 2015, 241: 177-188.
doi: 10.1016/j.plantsci.2015.10.011 pmid: 26706069 |
| [36] | Cai YH, Chen XJ, Xie K, et al. Dlf1, a WRKY transcription factor, is involved in the control of flowering time and plant height in rice[J]. PLoS One, 2014, 9(7): e102529. |
| [37] |
Shehzad T, Okuno K. QTL mapping for yield and yield-contributing traits in sorghum(Sorghum bicolor(L.)Moench)with genome-based SSR markers[J]. Euphytica, 2015, 203(1): 17-31.
doi: 10.1007/s10681-014-1243-9 URL |
| [17] | 陆平. 高粱种质资源描述规范和数据标准[M]. 北京: 中国农业出版社, 2006. |
| Lu P. Descriptors and data standard for sorghum[Sorghum bicolor(L.) Moench][M]. Beijing: China Agriculture Press, 2006. | |
| [18] |
曹永策, 李曙光, 张新草, 等. 夏大豆重组自交系群体遗传图谱构建及开花期QTL分析[J]. 中国农业科学, 2020, 53(4): 683-694.
doi: 10.3864/j.issn.0578-1752.2020.04.002 |
|
Cao YC, Li SG, Zhang XC, et al. Construction of genetic map and mapping QTL for flowering time in a summer planting soybean recombinant inbred line population[J]. Sci Agric Sin, 2020, 53(4): 683-694.
doi: 10.3864/j.issn.0578-1752.2020.04.002 |
|
| [19] |
Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA[J]. Nucleic Acids Res, 1980, 8(19): 4321-4325.
doi: 10.1093/nar/8.19.4321 pmid: 7433111 |
| [20] |
Qi P, Gimode D, Saha D, et al. UGbS-Flex, a novel bioinformatics pipeline for imputation-free SNP discovery in polyploids without a reference genome: finger millet as a case study[J]. BMC Plant Biol, 2018, 18(1): 117.
doi: 10.1186/s12870-018-1316-3 pmid: 29902967 |
| [21] |
丁延庆, 徐建霞, 汪灿, 等. 基于Super-GBS技术的高粱籽粒酿造相关性状QTL定位[J]. 核农学报, 2023, 37(2): 241-250.
doi: 10.11869/j.issn.1000-8551.2023.02.0241 |
| [38] |
林泽川, 曹立勇. 水稻株型相关基因的定位与克隆研究进展[J]. 中国稻米, 2014, 20(1): 17-22, 27.
doi: 10.3969/j.issn.1006-8082.2014.01.004 |
| Lin ZC, Cao LY. Progress on mapping and cloning of genes related to rice plant type[J]. China Rice, 2014, 20(1): 17-22, 27. | |
| [39] |
王文秀, 王磊. 玉米矮杆基因研究进展[J]. 生物技术通报, 2018, 34(11): 22-26.
doi: 10.13560/j.cnki.biotech.bull.1985.2018-0444 URL |
| Wang WX, Wang L. Research progress on maize dwarf genes[J]. Biotechnol Bull, 2018, 34(11): 22-26. | |
| [40] |
吕广德, 靳雪梅, 郭营, 等. 小麦株高分子遗传学研究进展[J]. 植物遗传资源学报, 2021, 22(3): 571-582.
doi: 10.13430/j.cnki.jpgr.20200927001 |
|
Lyu GD, Jin XM, Guo Y, et al. Advances in molecular genetics of wheat plant height[J]. J Plant Genet Resour, 2021, 22(3): 571-582.
doi: 10.13430/j.cnki.jpgr.20200927001 |
|
| [41] | Ge FY, Xie P, Wu YR, et al. Genetic architecture and molecular regulation of sorghum domestication[J]. aBIOTECH, 2022: 1-15. |
| [42] |
Hao HQ, Li ZG, Leng CY, et al. sorghum breeding in the genomic era: opportunities and challenges[J]. Theor Appl Genet, 2021, 134(7): 1899-1924.
doi: 10.1007/s00122-021-03789-z pmid: 33655424 |
| [1] | 王宝宝, 王海洋. 理想株型塑造之于玉米耐密改良[J]. 生物技术通报, 2023, 39(8): 11-30. |
| [2] | 吴元明, 林佳怡, 柳雨汐, 李丹婷, 张宗琼, 郑晓明, 逄洪波. 基于BSA-seq和RNA-seq挖掘水稻株高相关QTL[J]. 生物技术通报, 2023, 39(8): 173-184. |
| [3] | 方澜, 黎妍妍, 江健伟, 成胜, 孙正祥, 周燚. 盘龙参内生真菌胞内细菌7-2H的分离鉴定和促生特性研究[J]. 生物技术通报, 2023, 39(8): 272-282. |
| [4] | 范昕琦, 王海燕, 陈静, 张晓娟, 郭琦, 梁笃, 周福平, 聂萌恩, 张一中, 柳青山. EMS诱变对高粱成苗及M1主要农艺性状的影响[J]. 生物技术通报, 2023, 39(7): 173-184. |
| [5] | 王春语, 李政君, 王平, 张丽霞. 高粱表皮蜡质缺失突变体sb1抗旱生理生化分析[J]. 生物技术通报, 2023, 39(5): 160-167. |
| [6] | 黄婧, 朱亮, 薛蓬勃, 付强. 水稻叶和籽粒镉积累机制及QTL定位研究[J]. 生物技术通报, 2022, 38(8): 118-126. |
| [7] | 周诗晨, 仪治本, 王馨翊, 杨晓颖, 孙丽娜, 栾维江, 梁闪闪. 高粱双粒突变体Dgs的遗传分析与基因定位[J]. 生物技术通报, 2022, 38(7): 171-177. |
| [8] | 杨益善, 孙平勇, 余木兰. 水稻不育系稻粒黑粉病抗性QTL的定位[J]. 生物技术通报, 2022, 38(3): 16-21. |
| [9] | 江佰阳, 白文斌, 张建华, 范娜, 史丽娟. 高粱抗旱性鉴定方法及分子生物学研究进展[J]. 生物技术通报, 2021, 37(4): 260-272. |
| [10] | 张一中, 范昕琦, 杨慧勇, 张晓娟, 邵强, 梁笃, 郭琦, 柳青山, 杜维俊. 基于简化基因组测序高粱育种材料亲缘关系的分析[J]. 生物技术通报, 2020, 36(12): 21-33. |
| [11] | 张丹, 王楠, 李超, 谢旗, 唐三元. 甜高粱——一种优质的饲料作物[J]. 生物技术通报, 2019, 35(5): 2-8. |
| [12] | 冷传远, 郝怀庆, 景海春. 甜高粱茎秆持汁性研究进展[J]. 生物技术通报, 2019, 35(5): 9-14. |
| [13] | 韩立杰, 才宏伟. 高粱粒重遗传研究进展[J]. 生物技术通报, 2019, 35(5): 15-27. |
| [14] | 丁延庆, 周棱波, 汪灿, 曹宁, 程斌, 高旭, 彭秋, 邵明波, 张立异. 酱香型酒用糯高粱研究进展[J]. 生物技术通报, 2019, 35(5): 28-34. |
| [15] | 宋玉双, 隋娜. 甜高粱FAD7基因的功能分析[J]. 生物技术通报, 2019, 35(5): 35-41. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||