








生物技术通报 ›› 2024, Vol. 40 ›› Issue (3): 181-192.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0960
梅显军1,2( ), 宋慧洋2, 李京昊2, 梅超2,3, 宋倩娜2,3, 冯瑞云2,3(
), 宋慧洋2, 李京昊2, 梅超2,3, 宋倩娜2,3, 冯瑞云2,3( ), 陈喜明1,2(
), 陈喜明1,2( )
)
收稿日期:2023-10-11
出版日期:2024-03-26
发布日期:2024-04-08
通讯作者:
陈喜明,男,硕士,研究员,研究方向:杂粮育种与高产栽培;E-mail: 516834898@qq.com;作者简介:梅显军,男,硕士研究生,研究方向:杂粮遗传育种;E-mail: 1732244325@qq.com
基金资助:
MEI Xian-jun1,2( ), SONG Hui-yang2, LI Jing-hao2, MEI Chao2,3, SONG Qian-na2,3, FENG Rui-yun2,3(
), SONG Hui-yang2, LI Jing-hao2, MEI Chao2,3, SONG Qian-na2,3, FENG Rui-yun2,3( ), CHEN Xi-ming1,2(
), CHEN Xi-ming1,2( )
)
Received:2023-10-11
Published:2024-03-26
Online:2024-04-08
摘要:
【目的】 DNA binding with one finger(Dof)基因家族是植物特有的转录因子在植物生长发育和胁迫响应中发挥重要作用。研究Dof转录因子在马铃薯响应逆境胁迫特别是盐胁迫中的功能与机制,为获得抗盐逆境胁迫新品种提供一定的理论和技术基础。【方法】 克隆StDof5,并对其进行生物信息学分析,通过同源重组的方法构建StDof5过表达载体,转化马铃薯得到阳性植株,并对其表达进行分析。【结果】 StDof5的CDS全长498 bp,编码165个氨基酸,分子式为C800H1236N242O246S9,StDof5原子个数2 533个,相对分子量为18 468.63 kD,等电点是8.47,脂肪族指数为51.45,不稳定系数为45.16,预测该蛋白为不稳定蛋白。预测得到的跨膜螺旋个数为0,其属于非跨膜蛋白。StDof5蛋白亲水性平均值为-0.876,预测该蛋白属于亲水性蛋白。StDof5蛋白二级结构预测结果显示α螺旋占20%,β转角占0,延长链占9.7%,无规卷曲占66.06%。盐胁迫表型试验表明,过表达植株(OPs)比野生型(DES)的株高、鲜重均有明显程度上升,但根长、根数和叶片数差异不大。【结论】 过表达植株(OPs)能够抵御盐胁迫,对盐胁迫呈现脱敏感表型。该基因对盐胁迫响应明显,且存在组织特异性。
梅显军, 宋慧洋, 李京昊, 梅超, 宋倩娜, 冯瑞云, 陈喜明. 马铃薯StDof5的克隆及表达分析[J]. 生物技术通报, 2024, 40(3): 181-192.
MEI Xian-jun, SONG Hui-yang, LI Jing-hao, MEI Chao, SONG Qian-na, FENG Rui-yun, CHEN Xi-ming. Cloning and Expression Analysis of StDof5 Gene in Potato[J]. Biotechnology Bulletin, 2024, 40(3): 181-192.
| 引物名称Primer name | 引物序列Primer sequence(5'-3') |
|---|---|
| Actin-F | GGGATGGAGAAGTTTGGTGGTGG |
| Actin-R | CTTCGACCAAGGGATGGTGTAGC |
| Dof-F.2 | GCTTGACTTTGATGGAGTGGTGG |
| Dof-R.2 | AAGATTGGCTATCGGAGGTGC |
| ABF4-F | CTTCTGCACATACAAGGATATTCAA |
| ABF4-R | TGCCTAGCCAAAGGGAAATCA |
| MYB2-F | ATGACATGGACCTCCGACGA |
| MYB2-R | CGTTCGCTTTAGACCAGCAC |
| SnRK2s-F | TTGCTGTAGAGAAGGTGGGT |
| SnRK2s-R | GTCCTACTGTCACTGCCGTC |
| DREB2A-F | CACAGAAGAAAAGAGAAATGGCT |
| DREB2A-R | TCCACTATAATCCAACGGCAGA |
| ABI3-F | TGCTGACATGAAGTGTGGCA |
| ABI3-R | TTTTGCCCTCCGACTTTGGT |
| ABI5-F | TAAAACAGGCCCTGGCGGAG |
| ABI5-R | TTGCGCCTTTGTTTGAGCTT |
表1 荧光定量引物序列
Table 1 Primer sequences for fluorescence quantification
| 引物名称Primer name | 引物序列Primer sequence(5'-3') |
|---|---|
| Actin-F | GGGATGGAGAAGTTTGGTGGTGG |
| Actin-R | CTTCGACCAAGGGATGGTGTAGC |
| Dof-F.2 | GCTTGACTTTGATGGAGTGGTGG |
| Dof-R.2 | AAGATTGGCTATCGGAGGTGC |
| ABF4-F | CTTCTGCACATACAAGGATATTCAA |
| ABF4-R | TGCCTAGCCAAAGGGAAATCA |
| MYB2-F | ATGACATGGACCTCCGACGA |
| MYB2-R | CGTTCGCTTTAGACCAGCAC |
| SnRK2s-F | TTGCTGTAGAGAAGGTGGGT |
| SnRK2s-R | GTCCTACTGTCACTGCCGTC |
| DREB2A-F | CACAGAAGAAAAGAGAAATGGCT |
| DREB2A-R | TCCACTATAATCCAACGGCAGA |
| ABI3-F | TGCTGACATGAAGTGTGGCA |
| ABI3-R | TTTTGCCCTCCGACTTTGGT |
| ABI5-F | TAAAACAGGCCCTGGCGGAG |
| ABI5-R | TTGCGCCTTTGTTTGAGCTT |

图1 StDof5蛋白生物信息学分析 A:StDof5蛋白的跨膜结构域;B:马铃薯 StDof5蛋白的二级结构预测;C:马铃薯StDof5蛋白质的亲水性;D:马铃薯 StDof5蛋白的三级结构预测
Fig. 1 Bioinformatics analysis of StDof5 protein A: Transmembrane domain of the StDof5 protein; B: secondary structure prediction of potato StDof5 protein; C: hydrophilic nature of potato StDof5 protein; D: tertiary structure prediction of potato StDof5 protein
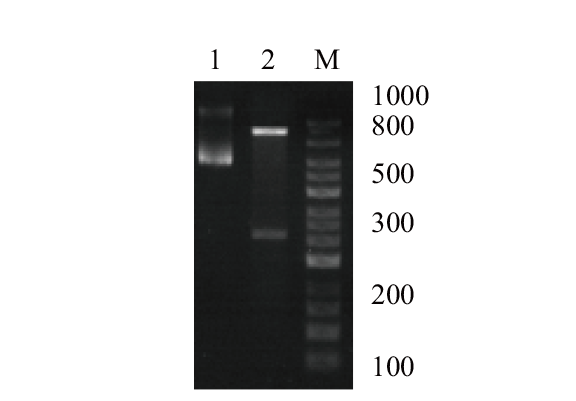
图4 马铃薯StDof5的PCR扩增 1:含目的基因StDof5的质粒;2:Hind III-Sac I酶切质粒;M:1 kb marker
Fig. 4 PCR amplification of the potato StDof5 gene 1: Plasmid containing target gene StDof5. 2: Hind III-Sac I digested plasmid. M: 1 kb marker
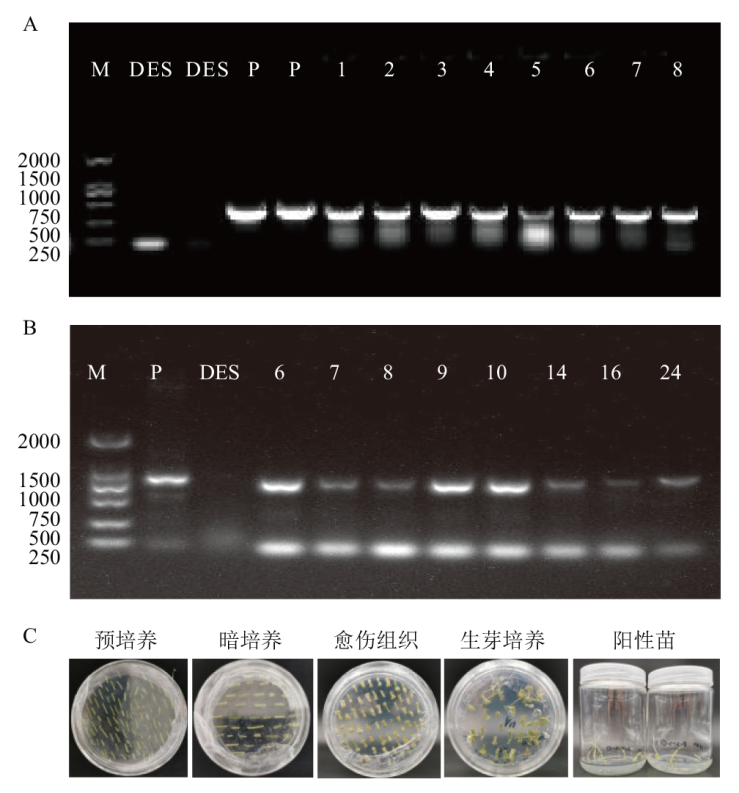
图5 马铃薯StDof5载体的构建 A:农杆菌载体构建;B:过表达马铃薯植株鉴定;C:马铃薯遗传转化组织培养;M: marker 2000 bp;P:质粒;DES:野生型,1-8:阳性单克隆;6-10、14、16、24:过表达阳性植株
Fig. 5 Construction of the potato StDof5 vector A: Agrobacterium vector construction; B: potato genetically transformed tissue culture; C: identification of overexpressed potato plants. M: Marker 2000 bp. P: Plasmid. DES: Wild type. 1-8: Positive single clone. 6-10, 14, 16, 24: Overexpression positive plants
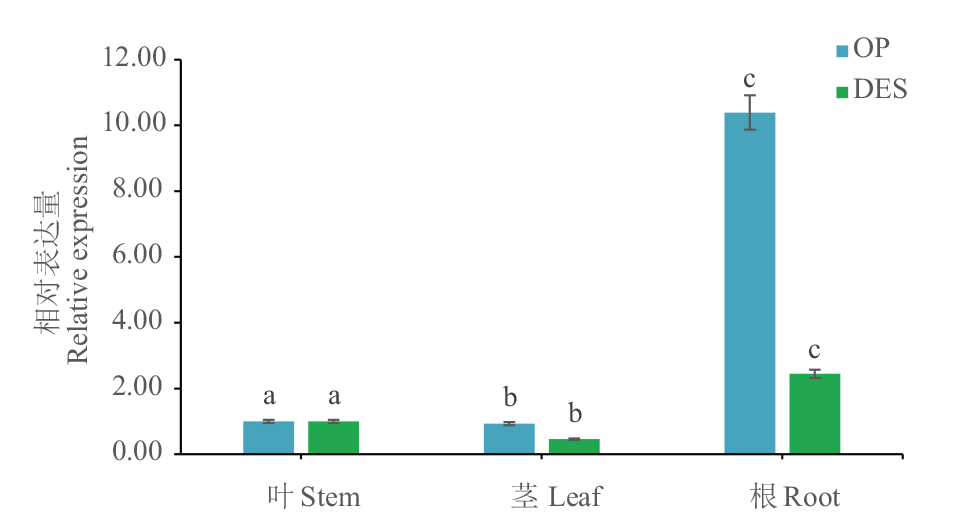
图6 不同组织中马铃薯Dof5的表达量分析 DES:野生型;OP:过表达阳性植株;不同字母表示差异显著(P<0.05)。下同
Fig. 6 Analysis of Dof5 gene expression in different tissues of potato DES: Wild type. OP: Overexpressed positive plants. Different letters indicate a significant difference(P<0.05). The same below
| 测量指标Measurement index | 盐浓度Salting strength/(mol·L-1) | 品种名称Variety name | ||||||
|---|---|---|---|---|---|---|---|---|
| DES | OP-6 | OP-7 | OP-9 | OP-10 | OP-16 | OP-24 | ||
| 株高Plant height/cm | 0 | 6.87±0.38a | 7.10±0.17a | 5.57±0.40a | 5.43±0.15a | 6.53±0.85a | 7.37±0.75a | 7.83±1.01a |
| 50 | 3.70±010b | 4.83±0.65b | 3.70±0.36b | 4.37±0.21b | 3.97±0.35b | 6.00±0.72b | 4.13±0.45b | |
| 100 | 2.13±0.76c | 2.97±0.42c | 2.00±0.53c | 1.77±0.31c | 2.80±0.36c | 3.00±0.44c | 2.93±0.15c | |
| 鲜重Fresh weight/g | 0 | 0.21±0.01a | 0.23±0.01a | 0.19±0.01a | 0.21±0.02a | 0.24±0.02a | 0.19±0.02a | 0.35±0.01a |
| 50 | 0.14±0.01b | 0.19±0.02b | 0.12±0.02b | 0.16±0.02b | 0.16±0.02b | 0.16±0.01a | 0.28±0.02b | |
| 100 | 0.09±0.01c | 0.12±0.02c | 0.09±0.01c | 0.09±0.01c | 0.12±0.01c | 0.12±0.02b | 0.15±0.02c | |
| 根长Root length/cm | 0 | 12.13±0.25a | 12.20±0.46a | 12.23±2.03a | 14.70±0.62a | 17.63±0.35a | 12.90±1.49a | 12.03±0.72a |
| 50 | 9.00±0.26b | 9.73±0.12b | 10.93±1.79ab | 13.07±0.31b | 13.73±0.31b | 10.53±0.61b | 11.67±0.31a | |
| 100 | 7.53±0.25c | 8.93±060b | 6.70±2.74b | 4.00±0.72c | 10.1±0.20c | 9.73±0.31b | 8.60±0.53b | |
| 根数Root | 0 | 6.33±1.15a | 7.33 ±2.31a | 8.00±0.00a | 7.00±1.00a | 8.67±1.53a | 6.00±0.00a | 9.67±2.08a |
| 50 | 3.67±2.08a | 5.67±2.31a | 4.00±1.00b | 5.33±2.31a | 4.67±1.53b | 6.67±2.89a | 5.67±1.15b | |
| 100 | 3.33±1.53a | 5.00±1.00a | 3.33±2.08b | 1.00±0.00b | 4.33±2.31b | 1.67±0.58b | 3.00±1.73b | |
| 叶片数Number of leaves | 0 | 7.33±0.58a | 8.33±0.58a | 7.67±0.58a | 6.67±0.58a | 7.00±0.00a | 7.00±1.00a | 6.67±0.58a |
| 50 | 6.33±0.58a | 7.00±0.00a | 6.33±1.15a | 6.00±1.00ab | 7.00±0.00a | 7.00±1.00a | 6.66±0.58a | |
| 100 | 7.00±1.00a | 7.00±1.00a | 6.67±0.58a | 5.00±0.00b | 6.33±1.15a | 6.00±1.00a | 6.67±0.58a | |
表2 盐胁迫生长指标的测定
Table 2 Determination of the growth indicators of salt stress
| 测量指标Measurement index | 盐浓度Salting strength/(mol·L-1) | 品种名称Variety name | ||||||
|---|---|---|---|---|---|---|---|---|
| DES | OP-6 | OP-7 | OP-9 | OP-10 | OP-16 | OP-24 | ||
| 株高Plant height/cm | 0 | 6.87±0.38a | 7.10±0.17a | 5.57±0.40a | 5.43±0.15a | 6.53±0.85a | 7.37±0.75a | 7.83±1.01a |
| 50 | 3.70±010b | 4.83±0.65b | 3.70±0.36b | 4.37±0.21b | 3.97±0.35b | 6.00±0.72b | 4.13±0.45b | |
| 100 | 2.13±0.76c | 2.97±0.42c | 2.00±0.53c | 1.77±0.31c | 2.80±0.36c | 3.00±0.44c | 2.93±0.15c | |
| 鲜重Fresh weight/g | 0 | 0.21±0.01a | 0.23±0.01a | 0.19±0.01a | 0.21±0.02a | 0.24±0.02a | 0.19±0.02a | 0.35±0.01a |
| 50 | 0.14±0.01b | 0.19±0.02b | 0.12±0.02b | 0.16±0.02b | 0.16±0.02b | 0.16±0.01a | 0.28±0.02b | |
| 100 | 0.09±0.01c | 0.12±0.02c | 0.09±0.01c | 0.09±0.01c | 0.12±0.01c | 0.12±0.02b | 0.15±0.02c | |
| 根长Root length/cm | 0 | 12.13±0.25a | 12.20±0.46a | 12.23±2.03a | 14.70±0.62a | 17.63±0.35a | 12.90±1.49a | 12.03±0.72a |
| 50 | 9.00±0.26b | 9.73±0.12b | 10.93±1.79ab | 13.07±0.31b | 13.73±0.31b | 10.53±0.61b | 11.67±0.31a | |
| 100 | 7.53±0.25c | 8.93±060b | 6.70±2.74b | 4.00±0.72c | 10.1±0.20c | 9.73±0.31b | 8.60±0.53b | |
| 根数Root | 0 | 6.33±1.15a | 7.33 ±2.31a | 8.00±0.00a | 7.00±1.00a | 8.67±1.53a | 6.00±0.00a | 9.67±2.08a |
| 50 | 3.67±2.08a | 5.67±2.31a | 4.00±1.00b | 5.33±2.31a | 4.67±1.53b | 6.67±2.89a | 5.67±1.15b | |
| 100 | 3.33±1.53a | 5.00±1.00a | 3.33±2.08b | 1.00±0.00b | 4.33±2.31b | 1.67±0.58b | 3.00±1.73b | |
| 叶片数Number of leaves | 0 | 7.33±0.58a | 8.33±0.58a | 7.67±0.58a | 6.67±0.58a | 7.00±0.00a | 7.00±1.00a | 6.67±0.58a |
| 50 | 6.33±0.58a | 7.00±0.00a | 6.33±1.15a | 6.00±1.00ab | 7.00±0.00a | 7.00±1.00a | 6.66±0.58a | |
| 100 | 7.00±1.00a | 7.00±1.00a | 6.67±0.58a | 5.00±0.00b | 6.33±1.15a | 6.00±1.00a | 6.67±0.58a | |

图9 盐胁迫25 d后组培苗生长状态 图片从上到下依次是:DES、OP-6、OP-7、OP-9、OP-10、OP-16、OP-24,图片内分别为梯度0、50和100 mmol/L NaCl浓度的盐胁迫
Fig. 9 Growth status of tissue culture seedlings after 25 d of salt stress From top to bottom, the pictures are: DES, OP-6, OP-7, OP-9, OP-10, OP-16, OP-24, with the salt stress of 0, 50 and 100 mmol/L NaCl, respectively
| [1] |
Gupta S, Malviya N, Kushwaha H, et al. Insights into structural and functional diversity of Dof(DNA binding with one finger)transcription factor[J]. Planta, 2015, 241(3): 549-562.
doi: 10.1007/s00425-014-2239-3 pmid: 25564353 |
| [2] |
Washio K. Functional dissections between GAMYB and Dof transcription factors suggest a role for protein-protein associations in the gibberellin-mediated expression of the RAmy1A gene in the rice aleurone[J]. Plant Physiol, 2003, 133(2): 850-863.
pmid: 14500792 |
| [3] |
Wang CM, Song BB, Dai YQ, et al. Genome-wide identification and functional analysis of U-box E3 ubiquitin ligases gene family related to drought stress response in Chinese white pear(Pyrus bretschneideri)[J]. BMC Plant Biol, 2021, 21(1): 235.
doi: 10.1186/s12870-021-03024-3 |
| [4] |
Wang T, Yue JJ, Wang XJ, et al. Genome-wide identification and characterization of the Dof gene family in moso bamboo(Phyllostachys heterocycla var. pubescens)[J]. Genes Genom, 2016, 38(8): 733-745.
doi: 10.1007/s13258-016-0418-2 URL |
| [5] | Li Y, Ding WJ, Jiang HY, et al. Advances in research of the Dof gene family in plant[J]. Acta Botanica Boreali-Occidentalia Sinica, 2018, 38(9): 1758-1766. |
| [6] |
Yanagisawa S, Schmidt RJ. Diversity and similarity among recognition sequences of Dof transcription factors[J]. Plant J, 1999, 17(2): 209-214.
doi: 10.1046/j.1365-313x.1999.00363.x pmid: 10074718 |
| [7] |
Yanagisawa S. Dof domain proteins: Plant-specific transcription factors associated with diverse phenomena unique to plants[J]. Plant Cell Physiol, 2004, 45(4): 386-391.
doi: 10.1093/pcp/pch055 pmid: 15111712 |
| [8] |
Venkatesh J, Park SW. Genome-wide analysis and expression profiling of DNA-binding with one zinc finger(Dof)transcription factor family in potato[J]. Plant Physiol Biochem, 2015, 94: 73-85.
doi: 10.1016/j.plaphy.2015.05.010 URL |
| [9] |
Yanagisawa S, Izui K. Molecular cloning of two DNA-binding proteins of maize that are structurally different but interact with the same sequence motif[J]. J Biol Chem, 1993, 268(21): 16028-16036.
pmid: 8340424 |
| [10] | 张华珍, 吴昊, 李香花, 等. 一个低氮诱导表达的水稻Dof转录因子OsDof-13的分离和转化[J]. 分子植物育种, 2007, 5(4): 455-460. |
| Zhang HZ, Wu H, Li XH, et al. Isolation and transformation of OsDof-13, a member of rice Dof transcription factor family, induced by low nitrogen stress[J]. Mol Plant Breed, 2007, 5(4): 455-460. | |
| [11] |
Lijavetzky D, Carbonero P, Vicente-Carbajosa J. Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families[J]. BMC Evol Biol, 2003, 3: 17.
pmid: 12877745 |
| [12] |
Cai XF, Zhang YY, Zhang CJ, et al. Genome-wide analysis of plant-specific Dof transcription factor family in tomato[J]. J Integr Plant Biol, 2013, 55(6): 552-566.
doi: 10.1111/jipb.v55.6 URL |
| [13] | 马凯恒, 张彤, 闫鹏宇, 等. 核桃转录因子JrDof3的克隆及渗透胁迫表达分析[J]. 中南林业科技大学学报, 2020, 40(10): 35-41. |
| Ma KH, Zhang T, Yan PY, et al. Cloning and expression analysis of walnut transcription factor Dof3 to osmotic stress[J]. J Central South Univ For Technol, 2020, 40(10): 35-41. | |
| [14] |
Corrales AR, Nebauer SG, Carrillo L, et al. Characterization of tomato cycling Dof factors reveals conserved and new functions in the control of flowering time and abiotic stress responses[J]. J Exp Bot, 2014, 65(4): 995-1012.
doi: 10.1093/jxb/ert451 URL |
| [15] | 成亮, 刘宝玲, 刘盼, 等. 青狗尾草Dof基因家族的全基因组鉴定及组织特异性表达分析[J]. 分子植物育种, 2018, 16(13): 4226-4234. |
| Cheng L, Liu BL, Liu P, et al. Genome-wide analysis and expression profiling of dof family in Setaria viridis[J]. Mol Plant Breed, 2018, 16(13): 4226-4234. | |
| [16] |
Choi HI, Hong JH, Ha JO, et al. ABFs, a family of ABA-responsive element binding factors[J]. J Biol Chem, 2000, 275(3): 1723-1730.
doi: 10.1074/jbc.275.3.1723 pmid: 10636868 |
| [17] | 李杨. 玉米中PP2Cs与SnRK2s相互作用研究[D]. 合肥: 安徽农业大学, 2021. |
| Li Y. Study on the interaction between PP2Cs and SnRK2s in maize[D]. Hefei: Anhui Agricultural University, 2021. | |
| [18] | 赵芝婧. 拟南芥转录因子LFY2和MYB2在花药发育调控网络中的作用研究[D]. 北京林业大学, 2020. |
| Zhao ZJ. Role of Arabidopsis transcription factors LFY2 and MYB2 in anther development regulation network[D]. Beijing: Beijing Forestry University, 2020. | |
| [19] |
Achard P, Gong F, Cheminant S, et al. The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism[J]. Plant cell, 2008, 20(8): 2117-2129.
doi: 10.1105/tpc.108.058941 pmid: 18757556 |
| [20] |
Fujita Y, Fujita AM, Satoh CR, et al. AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis[J]. Plant Cell, 2005, 17(12): 3470-3488.
doi: 10.1105/tpc.105.035659 URL |
| [21] |
Zheng ZF, Xu XP, Crosley RA, et al. The protein kinase SnRK2.6 mediates the regulation of sucrose metabolism and plant growth in Arabidopsis[J]. Plant Physiol, 2010, 153(1): 99-113.
doi: 10.1104/pp.109.150789 URL |
| [22] |
Wang YP, Li L, Ye TT, et al. The inhibitory effect of ABA on floral transition is mediated by ABI5 in Arabidopsis[J]. J Exp Bot, 2013, 64(2): 675-684.
doi: 10.1093/jxb/ers361 URL |
| [23] | 王伟, 李立芹, 邹雪, 等. 马铃薯块茎总RNA提取方法比较[J]. 江苏农业科学, 2013, 41(1): 38-40. |
| Wang W, Li LQ, Zou X, et al. Comparison of extraction methods of total RNA from potato tuber[J]. Jiangsu Agric Sci, 2013, 41(1): 38-40. | |
| [24] | 于惠琳, 吴玉群, 王延波, 等. 甜玉米籽粒总RNA提取方法的比较[J]. 园艺与种苗, 2022, 42(1): 67-68, 71. |
| Yu HL, Wu YQ, Wang YB, et al. Comparison of total RNA extraction methods from sweet corn kernels[J]. Hortic Seed, 2022, 42(1): 67-68, 71. | |
| [25] | 张晓丽, 代红军. 植物RNA提取方法的研究进展[J]. 北方园艺, 2014(8): 175-178. |
| Zhang XL, Dai HJ. Research progress on extraction method of plant RNA[J]. North Hortic, 2014(8): 175-178. | |
| [26] | 赵勤. 农杆菌介导的马铃薯遗传转化影响因素研究进展[J]. 园艺与种苗, 2011, 31(2): 92-94. |
| Zhao Q. Study on influencing factors of Agrobacterium-mediated genetic transformation in potato[J]. Rain Fed Crops, 2011, 31(2): 92-94. | |
| [27] | 杨永智. 农杆菌介导法马铃薯遗传转化体系的优化[J]. 江苏农业学报, 2013, 29(4): 738-742. |
| Yang YZ. Optimization of Agrobacterium tumerfaciens mediated potato transformation system[J]. Jiangsu J Agric Sci, 2013, 29(4): 738-742. | |
| [28] | 连欢欢, 杨永智, 王舰. 根癌农杆菌介导的马铃薯DM茎段遗传转化条件的优化[J]. 分子植物育种, 2016, 14(6): 1491-1499. |
| Lian HH, Yang YZ, Wang J. Optimization of conditions for genetic transformation of potato DM stems[J]. Molecular plant breeding, 2016, 14(6): 1491-1499. | |
| [29] | 张萍. 根癌农杆菌介导的马铃薯遗传转化体系的研究[J]. 陕西农业科学, 2019, 65(4): 27-29, 40. |
| Zhang P. Study on genetic transformation system of potato mediated by Agrobacterium tumefaciens[J]. Shaanxi J Agric Sci, 2019, 65(4): 27-29, 40. | |
| [30] | 李逸菲, 王瑞博, 夏惠婷, 等. 农杆菌介导四季秋海棠遗传转化体系建立[J]. 东北农业大学学报, 2023, 54(5): 19-27. |
| Li YF, Wang RB, Xia HT, et al. Establishment of Agrobacteri-um-mediated genetic transformation system for Begonia semperflo-rens[J]. J Northeast Agric Univ, 2023, 54(5): 19-27. | |
| [31] | 宋倩娜, 梅超, 霍利光, 等. 马铃薯品种‘并薯6号’遗传转化体系的建立[J]. 中国马铃薯, 2021, 35(5): 385-396. |
| Song QN, Mei C, Huo LG, et al. Establishment of the genetic transformation system for potato variety ‘Bingshu 6’[J]. Chin potato J, 2021, 35(5): 385-396. | |
| [32] | 高志鹏, 杜玉萧, 冯霞, 等. 鄂马铃薯3号遗传转化体系的构建与表达[J]. 湖北农业科学, 2022, 61(9): 162-167, 191. |
| Gao ZP, Du YX, Feng X, et al. Construction and expression of the # 3 genetic transformation system of Hubei potato No.3[J]. Hubei Agric Sci, 2022, 61(9): 162-167, 191. | |
| [33] | 陈兆贵, 林燕文, 李希陶, 等. 马铃薯植株再生和遗传转化技术研究进展[J]. 智慧农业导刊, 2023, 3(9): 12-15. |
| Chen ZG, Lin YW, Li XT, et al. Research progress of plant regeneration and genetic transformation of potatoes[J]. J Smart Agri, 2023, 3(9): 12-15. | |
| [34] |
公鑫, 王业红, 张剑峰, 等. 马铃薯RPP13基因RNAi载体的构建及遗传转化[J]. 华北农学报, 2023, 38(1): 178-183.
doi: 10.7668/hbnxb.20193453 |
| Gong X, Wang YH, Zhang JF, et al. Construction and genetic transformation of the RNAi vector of potato RPP 13 gene[J]. Acta Agric Boreali Sin, 2023, 38(1): 178-183. | |
| [35] | 万丽丽, 王转茸, 汤谧, 等. 甜瓜遗传转化技术的优化及DRs类基因的应用[J]. 江苏农业科学, 2023, 51(7): 49-58. |
| Wan LL, Wang ZR, Tang M, et al. Optimization of genetic transformation technology of melon and the application of DRs class genes[J]. Jiangsu Agric Sci, 2023, 51(7): 49-58. | |
| [36] |
Corrales AR, Carrillo L, Lasierra P, et al. Multifaceted role of cycling DOF factor 3(CDF3)in the regulation of flowering time and abiotic stress responses in Arabidopsis[J]. Plant Cell Environ, 2017, 40(5): 748-764.
doi: 10.1111/pce.v40.5 URL |
| [37] |
Xu PP, Chen HY, Ying L, et al. AtDOF5.4/OBP4, a DOF transcription factor gene that negatively regulates cell cycle progression and cell expansion in Arabidopsis thaliana[J]. Sci Rep, 2016, 6: 27705.
doi: 10.1038/srep27705 |
| [38] |
van Zelm E, Zhang YX, Testerink C. Salt tolerance mechanisms of plants[J]. Annu Rev Plant Bio, 2020, 71: 403-433.
doi: 10.1146/annurev-arplant-050718-100005 URL |
| [39] |
Su Y, Liang W, Liu ZJ, et al. Overexpression of GhDof1 improved salt and cold tolerance and seed oil content in Gossypium hirsutum[J]. J Plant Physiol, 2017, 218: 222-234.
doi: 10.1016/j.jplph.2017.07.017 URL |
| [40] |
Cai XF, Zhang CJ, Shu WB, et al. The transcription factor SlDof22 involved in ascorbate accumulation and salinity stress in tomato[J]. Biochem Biophys Res Commun, 2016, 474(4): 736-741.
doi: 10.1016/j.bbrc.2016.04.148 URL |
| [41] |
Kim J, Kim JH, Lyu JI, et al. New insights into the regulation of leaf senescence in Arabidopsis[J]. J Exp Bot, 2018, 69(4): 787-799.
doi: 10.1093/jxb/erx287 URL |
| [42] |
Lee BD, Ri KM, Young KM, et al. The F-box protein FKF1inhibits dimerization of COP1 in the control of photoperiodic flowering[J]. Nature Communications, 2018, 8(1): 2259.
doi: 10.1038/s41467-017-02476-2 |
| [43] |
Wen CL, Cheng Q, Zhao LQ, et al. Identification and characterisation of Dof transcription factors in the cucumber genome[J]. Sci Rep, 2016, 6: 23072.
doi: 10.1038/srep23072 |
| [44] |
Noguero M, Atif RM, OCHATT S, et al. The role of the DNA-binding One Zinc Finger(Dof)transcription factor family in plants[J]. Plant Sci, 2013, 209: 32-45.
doi: 10.1016/j.plantsci.2013.03.016 URL |
| [45] |
Zhang ZR, Yuan L, Liu X, et al. Evolution analysis of Dof transcription factor family and their expression in response to multiple abiotic stresses in Malus domestica[J]. Gene, 2018, 639: 137-148.
doi: 10.1016/j.gene.2017.09.039 URL |
| [46] |
Ewas M, Khames E, et al. The tomato Dof Daily Fluctuations 1, DESDF1 acts as flowering accelerator and protector against various stresses[J]. Sci Rep, 2017, 7(1): 10299.
doi: 10.1038/s41598-017-10399-7 |
| [47] |
Ma J, Li MY, Wang F, et al. Genome-wide analysis of Dof family transcription factors and their responses to abiotic stresses in Chinese cabbage[J]. BMC Genomics, 2015, 16(1): 33.
doi: 10.1186/s12864-015-1242-9 URL |
| [48] |
Kang Y, Yang X, Liu Y, et al. Integration of mRNA and miRNA analysis reveals the molecular mechanism of potato(Solanum tuberosum L.)response to alkali stress[J]. International Journal of Biological Macromolecules, 2021, 182: 938-949.
doi: 10.1016/j.ijbiomac.2021.04.094 URL |
| [1] | 张玉, 石磊, 巩檑, 聂峰杰, 杨江伟, 刘璇, 杨文静, 张国辉, 颉瑞霞, 张丽. 马铃薯WOX基因家族的鉴定及在离体再生和非生物胁迫中的表达分析[J]. 生物技术通报, 2024, 40(3): 170-180. |
| [2] | 刘雯锦, 马瑞, 刘升燕, 杨江伟, 张宁, 司怀军. 马铃薯StCIPK11的克隆及响应干旱胁迫分析[J]. 生物技术通报, 2023, 39(9): 147-155. |
| [3] | 黄文莉, 李香香, 周炆婷, 罗莎, 姚维嘉, 马杰, 张芬, 沈钰森, 顾宏辉, 王建升, 孙勃. 利用CRISPR/Cas9技术靶向编辑青花菜BoZDS[J]. 生物技术通报, 2023, 39(2): 80-87. |
| [4] | 安苗, 王彤彤, 付逸婷, 夏俊俊, 彭锁堂, 段永红. 52个马铃薯遗传多样性分析及SSR分子身份证构建[J]. 生物技术通报, 2023, 39(12): 136-147. |
| [5] | 周家燕, 邹建, 陈卫英, 吴一超, 陈奚潼, 王倩, 曾文静, 胡楠, 杨军. 植物多基因干扰载体体系构建与效用分析[J]. 生物技术通报, 2023, 39(1): 115-126. |
| [6] | 孙威, 张艳, 王聿晗, 徐僡, 徐小蓉, 鞠志刚. 马缨杜鹃Rd3GT1的克隆及对矮牵牛花色形成的影响[J]. 生物技术通报, 2022, 38(9): 198-206. |
| [7] | 陆新华, 孙德权, 张秀梅. 介孔硅纳米粒作为植物细胞转基因载体的研究[J]. 生物技术通报, 2022, 38(7): 194-204. |
| [8] | 于国红, 刘朋程, 李磊, 李明哲, 崔海英, 郝洪波, 郭安强. 不同基因型马铃薯对干旱胁迫的生理响应[J]. 生物技术通报, 2022, 38(5): 56-63. |
| [9] | 高蒙, 李富婷, 魏湛林, 张赛行, 白如仟, 尚轶, 马玲. 二倍体马铃薯中SCFSLF复合体的组分分析[J]. 生物技术通报, 2022, 38(4): 117-125. |
| [10] | 雷春霞, 李灿辉, 陈永坤, 龚明. 马铃薯块茎形成的生理生化基础和分子机制[J]. 生物技术通报, 2022, 38(4): 44-57. |
| [11] | 杨佳慧, 孙玉萍, 陆雅宁, 刘欢, 卢存福, 陈玉珍. 拟南芥AtTERT对大肠杆菌非生物胁迫抗性的影响[J]. 生物技术通报, 2022, 38(2): 1-9. |
| [12] | 冯建英, 李立芹, 鲁黎明. 马铃薯bHLH转录因子家族全基因组鉴定与表达分析[J]. 生物技术通报, 2022, 38(2): 21-33. |
| [13] | 刘萌萌, 韩立军, 刘宝玲, 薛金爱, 李润植. 陆地棉GhSDP1及其启动子的克隆与表达分析[J]. 生物技术通报, 2022, 38(2): 34-43. |
| [14] | 党瑗, 李维, 苗向, 修宇, 林善枝. 山杏油体蛋白基因PsOLE4克隆及其调控油脂累积功能分析[J]. 生物技术通报, 2022, 38(11): 151-161. |
| [15] | 胡子曜, 代培红, 刘超, 玛迪娜·木拉提, 王倩, 吾尕力汗·阿不都维力, 赵燚, 孙玲, 徐诗佳, 李月. 陆地棉小GTP结合蛋白基因GhROP3的克隆、表达及VIGS载体的构建[J]. 生物技术通报, 2021, 37(9): 106-113. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||