








生物技术通报 ›› 2022, Vol. 38 ›› Issue (2): 21-33.doi: 10.13560/j.cnki.biotech.bull.1985.2020-1327
收稿日期:2020-10-28
出版日期:2022-02-26
发布日期:2022-03-09
作者简介:冯建英,女,硕士研究生,研究方向:植物分子生物学;E-mail: 基金资助:
FENG Jian-ying( ), LI Li-qin, LU Li-ming(
), LI Li-qin, LU Li-ming( )
)
Received:2020-10-28
Published:2022-02-26
Online:2022-03-09
摘要:
对马铃薯bHLH转录因子进行全基因组鉴定与表达模式分析,为其生物学功能研究提供借鉴。基于马铃薯基因组数据库和Pfam数据库,运用HMMER对马铃薯bHLH家族成员进行全基因组鉴定,剔除冗余序列后,利用ExPASy和MEME软件对候选序列进行基本理化性质及保守元件分析,并使用MEGA-X软件进行聚类分析;用MG2C和TBtools软件分别绘制染色体定位图和表达模式图。结果表明,从马铃薯基因组数据库中共检索出108个bHLH转录因子,其氨基酸数目为62-694个,分子量为7 527.78-75 939.94 Da,理论等电点为4.55-10.40。这些bHLH转录因子分布在马铃薯的12条染色体上,均具有典型的HLH结构域,可划分为16个亚族。基因表达模式分析表明,马铃薯bHLH家族成员具有组织表达特异性,多数响应盐、甘露醇、生长素、脱落酸、赤霉素及热等胁迫。马铃薯全基因组包括108个高度保守的bHLH基因家族成员,其表达具有组织特异性,并且响应非生物胁迫的诱导。
冯建英, 李立芹, 鲁黎明. 马铃薯bHLH转录因子家族全基因组鉴定与表达分析[J]. 生物技术通报, 2022, 38(2): 21-33.
FENG Jian-ying, LI Li-qin, LU Li-ming. Genome-wide Identification and Expression Analysis of the bHLH Transcription Factor Family in Solanum tuberosum[J]. Biotechnology Bulletin, 2022, 38(2): 21-33.
| 基因 Gene | 上游引物 Forward primer(5'-3') | 下游引物 Reverse primer(5'-3') |
|---|---|---|
| StbHLH2 | ATGTTCACCGCCTTATTG | CTGATGAGATTAGAGCCACC |
| StbHLH14 | CTAAAGGTGGAAGTGGGGTA | TTGGGGGTTGTCAATAAAAT |
| StbHLH15 | ACAAGTTGCCCACCTACC | TGGAAGTTCTCACTGCCTCT |
| StbHLH34 | AAAGGAACCGCCGCAAAC | GCGTCTTGGCTTCTAAGGATT |
| StbHLH39 | ATCCTCTTGCTCGCCTGTT | GGAATAGTAGATTTTTGGGGCA |
| StbHLH53 | ACCTCCTGTTCCGATGAT | ACCAAAGTTCCCTCCACT |
| StbHLH57 | CATTGGATAAAGTCACCGATAC | CGAATCTACAGACGACCTCA |
| StbHLH59 | GGCATTTTTCACGGTTGCCT | CCGTCGTCACTTCCCTATCG |
| StbHLH66 | AGCACTTGTTCCTGGTTGTGA | AATGGTCCTTCCAAGCCACTC |
| StbHLH80 | CCAGATTCACCCCGTCGTTT | CCTTGTGCTCACGAAACACC |
| StbHLH90 | CCAGTTCCACTACACCGTCC | GCCGGAGGATTTGACGGAAT |
| StbHLH108 | GTCAAATCGAAGAACACGCGG | CTGCGGCTTTCAGGGATAAGT |
表1 引物序列
Table 1 Primer sequences
| 基因 Gene | 上游引物 Forward primer(5'-3') | 下游引物 Reverse primer(5'-3') |
|---|---|---|
| StbHLH2 | ATGTTCACCGCCTTATTG | CTGATGAGATTAGAGCCACC |
| StbHLH14 | CTAAAGGTGGAAGTGGGGTA | TTGGGGGTTGTCAATAAAAT |
| StbHLH15 | ACAAGTTGCCCACCTACC | TGGAAGTTCTCACTGCCTCT |
| StbHLH34 | AAAGGAACCGCCGCAAAC | GCGTCTTGGCTTCTAAGGATT |
| StbHLH39 | ATCCTCTTGCTCGCCTGTT | GGAATAGTAGATTTTTGGGGCA |
| StbHLH53 | ACCTCCTGTTCCGATGAT | ACCAAAGTTCCCTCCACT |
| StbHLH57 | CATTGGATAAAGTCACCGATAC | CGAATCTACAGACGACCTCA |
| StbHLH59 | GGCATTTTTCACGGTTGCCT | CCGTCGTCACTTCCCTATCG |
| StbHLH66 | AGCACTTGTTCCTGGTTGTGA | AATGGTCCTTCCAAGCCACTC |
| StbHLH80 | CCAGATTCACCCCGTCGTTT | CCTTGTGCTCACGAAACACC |
| StbHLH90 | CCAGTTCCACTACACCGTCC | GCCGGAGGATTTGACGGAAT |
| StbHLH108 | GTCAAATCGAAGAACACGCGG | CTGCGGCTTTCAGGGATAAGT |
| 基因 Gene | 基因ID Gene ID | 氨基酸数Number of amino acid | 分子量 Molecular weight/Da | 等电点 pI | 基因 Gene | 基因ID Gene ID | 氨基酸数Number of amino acid | 分子量 Molecular weight/Da | 等电点 pI | |
|---|---|---|---|---|---|---|---|---|---|---|
| StbHLH1 | PGSC0003DMT400095438 | 295 | 33 008.42 | 8.21 | StbHLH39 | PGSC0003DMT400036951 | 551 | 59 473.80 | 5.78 | |
| StbHLH2 | PGSC0003DMT400065029 | 397 | 43 515.43 | 6.16 | StbHLH40 | PGSC0003DMT400014656 | 276 | 31 123.90 | 5.76 | |
| StbHLH3 | PGSC0003DMT400004497 | 471 | 53 156.01 | 5.18 | StbHLH41 | PGSC0003DMT400006783 | 311 | 34 469.88 | 6.45 | |
| StbHLH4 | PGSC0003DMT400086863 | 393 | 44 510.14 | 6.01 | StbHLH42 | PGSC0003DMT400015377 | 203 | 22 928.51 | 9.69 | |
| StbHLH5 | PGSC0003DMT400066807 | 464 | 50 747.96 | 7.62 | StbHLH43 | PGSC0003DMT400060615 | 224 | 25 030.63 | 7.56 | |
| StbHLH6 | PGSC0003DMT400000652 | 598 | 66 369.87 | 6.86 | StbHLH44 | PGSC0003DMT400027293 | 235 | 27 571.74 | 8.82 | |
| StbHLH7 | PGSC0003DMT400047220 | 452 | 49 967.36 | 5.61 | StbHLH45 | PGSC0003DMT400005446 | 345 | 38 526.03 | 5.87 | |
| StbHLH8 | PGSC0003DMT400073574 | 337 | 38 301.38 | 5.14 | StbHLH46 | PGSC0003DMT400033589 | 321 | 35 634.69 | 4.55 | |
| StbHLH9 | PGSC0003DMT400013074 | 353 | 39 806.96 | 6.33 | StbHLH47 | PGSC0003DMT400024185 | 204 | 22 750.35 | 9.88 | |
| StbHLH10 | PGSC0003DMT400087143 | 333 | 36 913.79 | 5.28 | StbHLH48 | PGSC0003DMT400016360 | 338 | 37 784.66 | 4.66 | |
| StbHLH11 | PGSC0003DMT400066257 | 305 | 34 150.70 | 7.11 | StbHLH49 | PGSC0003DMT400012591 | 382 | 41 627.38 | 6.33 | |
| StbHLH12 | PGSC0003DMT400066259 | 307 | 34 354.03 | 6.67 | StbHLH50 | PGSC0003DMT400020534 | 314 | 34 098.61 | 5.32 | |
| StbHLH13 | PGSC0003DMT400066261 | 304 | 33 738.26 | 7.14 | StbHLH51 | PGSC0003DMT400009603 | 406 | 44 073.59 | 7.66 | |
| StbHLH14 | PGSC0003DMT400066264 | 307 | 34 179.83 | 7.12 | StbHLH52 | PGSC0003DMT400050572 | 321 | 35 928.50 | 5.11 | |
| StbHLH15 | PGSC0003DMT400001888 | 442 | 49 692.41 | 5.65 | StbHLH53 | PGSC0003DMT400050585 | 520 | 56 513.05 | 5.21 | |
| StbHLH16 | PGSC0003DMT400040021 | 303 | 34 207.55 | 6.54 | StbHLH54 | PGSC0003DMT400002155 | 312 | 34 997.42 | 6.00 | |
| StbHLH17 | PGSC0003DMT400026937 | 326 | 36 823.76 | 5.23 | StbHLH55 | PGSC0003DMT400033757 | 210 | 23 748.34 | 8.89 | |
| StbHLH18 | PGSC0003DMT400021383 | 244 | 27 458.61 | 5.56 | StbHLH56 | PGSC0003DMT400044959 | 231 | 26 357.79 | 5.64 | |
| StbHLH19 | PGSC0003DMT400026449 | 235 | 26 275.12 | 8.64 | StbHLH57 | PGSC0003DMT400018046 | 578 | 64 285.91 | 7.61 | |
| StbHLH20 | PGSC0003DMT400036021 | 442 | 49 217.42 | 5.90 | StbHLH58 | PGSC0003DMT400040761 | 390 | 44 069.48 | 5.27 | |
| StbHLH21 | PGSC0003DMT400055910 | 267 | 29 914.13 | 7.80 | StbHLH59 | PGSC0003DMT400038134 | 515 | 56 408.13 | 5.13 | |
| StbHLH22 | PGSC0003DMT400078776 | 317 | 35 038.85 | 6.12 | StbHLH60 | PGSC0003DMT400048941 | 312 | 35 241.59 | 4.72 | |
| StbHLH23 | PGSC0003DMT400078675 | 335 | 38 417.26 | 5.44 | StbHLH61 | PGSC0003DMT400040448 | 225 | 25 610.16 | 7.62 | |
| StbHLH24 | PGSC0003DMT400078696 | 347 | 39 529.70 | 6.39 | StbHLH62 | PGSC0003DMT400071861 | 62 | 7 527.78 | 10.4 | |
| StbHLH25 | PGSC0003DMT400078698 | 209 | 23 532.27 | 6.17 | StbHLH63 | PGSC0003DMT400080418 | 260 | 28 511.99 | 7.81 | |
| StbHLH26 | PGSC0003DMT400078699 | 323 | 35 931.15 | 5.47 | StbHLH64 | PGSC0003DMT400012487 | 390 | 42 992.79 | 5.76 | |
| StbHLH27 | PGSC0003DMT400010253 | 376 | 41 285.79 | 5.04 | StbHLH65 | PGSC0003DMT400074075 | 530 | 57 936.63 | 5.31 | |
| StbHLH28 | PGSC0003DMT400034838 | 296 | 32 476.68 | 5.27 | StbHLH66 | PGSC0003DMT400083164 | 416 | 46 807.69 | 8.71 | |
| StbHLH29 | PGSC0003DMT400012997 | 335 | 38 164.49 | 5.23 | StbHLH66 | PGSC0003DMT400069569 | 284 | 31 977.18 | 6.76 | |
| StbHLH30 | PGSC0003DMT400058169 | 268 | 30 772.88 | 7.63 | StbHLH68 | PGSC0003DMT400051980 | 467 | 51 763.96 | 7.19 | |
| StbHLH31 | PGSC0003DMT400048857 | 148 | 16 910.86 | 9.75 | StbHLH69 | PGSC0003DMT400051674 | 477 | 52 903.87 | 6.59 | |
| StbHLH32 | PGSC0003DMT400071293 | 224 | 25 544.12 | 8.60 | StbHLH70 | PGSC0003DMT400028917 | 444 | 46 271.20 | 6.09 | |
| StbHLH33 | PGSC0003DMT400019550 | 214 | 24 405.08 | 8.94 | StbHLH71 | PGSC0003DMT400010463 | 252 | 28 629.96 | 4.97 | |
| StbHLH34 | PGSC0003DMT400023497 | 355 | 38 301.12 | 5.94 | StbHLH72 | PGSC0003DMT400056685 | 374 | 42 308.55 | 8.80 | |
| StbHLH35 | PGSC0003DMT400051137 | 338 | 37 637.08 | 6.08 | StbHLH73 | PGSC0003DMT400041156 | 517 | 57 437.21 | 7.09 | |
| StbHLH36 | PGSC0003DMT400062844 | 291 | 30 828.06 | 5.65 | StbHLH74 | PGSC0003DMT400067287 | 351 | 39 693.02 | 8.14 | |
| StbHLH37 | PGSC0003DMT400063026 | 536 | 58 417.99 | 7.35 | StbHLH75 | PGSC0003DMT400052622 | 255 | 27 880.55 | 6.92 | |
| StbHLH38 | PGSC0003DMT400063117 | 437 | 48 174.88 | 6.79 | StbHLH76 | PGSC0003DMT400018274 | 224 | 25 831.73 | 6.32 | |
| StbHLH77 | PGSC0003DMT400049504 | 485 | 52 745.68 | 6.07 | StbHLH93 | PGSC0003DMT400016793 | 348 | 36 847.65 | 8.57 | |
| StbHLH78 | PGSC0003DMT400002963 | 598 | 65 258.62 | 5.92 | StbHLH94 | PGSC0003DMT400064399 | 388 | 42 575.81 | 5.83 | |
| StbHLH79 | PGSC0003DMT400015017 | 456 | 51 791.64 | 6.22 | StbHLH95 | PGSC0003DMT400028057 | 389 | 44 514.29 | 5.88 | |
| StbHLH80 | PGSC0003DMT400021165 | 435 | 49 379.65 | 5.89 | StbHLH96 | PGSC0003DMT400008370 | 198 | 22 043.97 | 8.88 | |
| StbHLH81 | PGSC0003DMT400053651 | 319 | 36 510.05 | 4.85 | StbHLH97 | PGSC0003DMT400089728 | 305 | 34 320.40 | 5.48 | |
| StbHLH82 | PGSC0003DMT400095303 | 311 | 35 808.26 | 4.90 | StbHLH98 | PGSC0003DMT400076759 | 180 | 20 751.84 | 9.58 | |
| StbHLH83 | PGSC0003DMT400045210 | 330 | 36 732.34 | 6.10 | StbHLH99 | PGSC0003DMT400021130 | 360 | 39 675.21 | 5.92 | |
| StbHLH84 | PGSC0003DMT400045204 | 694 | 75 939.94 | 5.47 | StbHLH100 | PGSC0003DMT400021062 | 380 | 41 565.47 | 9.10 | |
| StbHLH85 | PGSC0003DMT400032139 | 360 | 40 669.07 | 6.81 | StbHLH101 | PGSC0003DMT400041644 | 237 | 27 276.91 | 5.27 | |
| StbHLH86 | PGSC0003DMT400031899 | 431 | 48 790.06 | 6.71 | StbHLH102 | PGSC0003DMT400073760 | 262 | 30 275.48 | 5.87 | |
| StbHLH87 | PGSC0003DMT400053639 | 492 | 53 992.33 | 7.13 | StbHLH103 | PGSC0003DMT400056716 | 441 | 48 896.06 | 6.65 | |
| StbHLH88 | PGSC0003DMT400048756 | 578 | 62 943.67 | 6.42 | StbHLH104 | PGSC0003DMT400020244 | 382 | 39 561.65 | 6.21 | |
| StbHLH89 | PGSC0003DMT400002512 | 284 | 31 495.30 | 5.46 | StbHLH105 | PGSC0003DMT400013766 | 272 | 31 224.18 | 6.76 | |
| StbHLH90 | PGSC0003DMT400033569 | 590 | 64 760.08 | 5.81 | StbHLH106 | PGSC0003DMT400045820 | 296 | 33 899.60 | 7.02 | |
| StbHLH91 | PGSC0003DMT400076223 | 285 | 32 950.42 | 6.27 | StbHLH107 | PGSC0003DMT400011651 | 413 | 46 698.70 | 5.92 | |
| StbHLH92 | PGSC0003DMT400042791 | 439 | 49 242.61 | 6.06 | StbHLH108 | PGSC0003DMT400061284 | 489 | 54 218.28 | 5.12 |
表2 基本理化性质分析
Table 2 Analysis of basic physical and chemical properties
| 基因 Gene | 基因ID Gene ID | 氨基酸数Number of amino acid | 分子量 Molecular weight/Da | 等电点 pI | 基因 Gene | 基因ID Gene ID | 氨基酸数Number of amino acid | 分子量 Molecular weight/Da | 等电点 pI | |
|---|---|---|---|---|---|---|---|---|---|---|
| StbHLH1 | PGSC0003DMT400095438 | 295 | 33 008.42 | 8.21 | StbHLH39 | PGSC0003DMT400036951 | 551 | 59 473.80 | 5.78 | |
| StbHLH2 | PGSC0003DMT400065029 | 397 | 43 515.43 | 6.16 | StbHLH40 | PGSC0003DMT400014656 | 276 | 31 123.90 | 5.76 | |
| StbHLH3 | PGSC0003DMT400004497 | 471 | 53 156.01 | 5.18 | StbHLH41 | PGSC0003DMT400006783 | 311 | 34 469.88 | 6.45 | |
| StbHLH4 | PGSC0003DMT400086863 | 393 | 44 510.14 | 6.01 | StbHLH42 | PGSC0003DMT400015377 | 203 | 22 928.51 | 9.69 | |
| StbHLH5 | PGSC0003DMT400066807 | 464 | 50 747.96 | 7.62 | StbHLH43 | PGSC0003DMT400060615 | 224 | 25 030.63 | 7.56 | |
| StbHLH6 | PGSC0003DMT400000652 | 598 | 66 369.87 | 6.86 | StbHLH44 | PGSC0003DMT400027293 | 235 | 27 571.74 | 8.82 | |
| StbHLH7 | PGSC0003DMT400047220 | 452 | 49 967.36 | 5.61 | StbHLH45 | PGSC0003DMT400005446 | 345 | 38 526.03 | 5.87 | |
| StbHLH8 | PGSC0003DMT400073574 | 337 | 38 301.38 | 5.14 | StbHLH46 | PGSC0003DMT400033589 | 321 | 35 634.69 | 4.55 | |
| StbHLH9 | PGSC0003DMT400013074 | 353 | 39 806.96 | 6.33 | StbHLH47 | PGSC0003DMT400024185 | 204 | 22 750.35 | 9.88 | |
| StbHLH10 | PGSC0003DMT400087143 | 333 | 36 913.79 | 5.28 | StbHLH48 | PGSC0003DMT400016360 | 338 | 37 784.66 | 4.66 | |
| StbHLH11 | PGSC0003DMT400066257 | 305 | 34 150.70 | 7.11 | StbHLH49 | PGSC0003DMT400012591 | 382 | 41 627.38 | 6.33 | |
| StbHLH12 | PGSC0003DMT400066259 | 307 | 34 354.03 | 6.67 | StbHLH50 | PGSC0003DMT400020534 | 314 | 34 098.61 | 5.32 | |
| StbHLH13 | PGSC0003DMT400066261 | 304 | 33 738.26 | 7.14 | StbHLH51 | PGSC0003DMT400009603 | 406 | 44 073.59 | 7.66 | |
| StbHLH14 | PGSC0003DMT400066264 | 307 | 34 179.83 | 7.12 | StbHLH52 | PGSC0003DMT400050572 | 321 | 35 928.50 | 5.11 | |
| StbHLH15 | PGSC0003DMT400001888 | 442 | 49 692.41 | 5.65 | StbHLH53 | PGSC0003DMT400050585 | 520 | 56 513.05 | 5.21 | |
| StbHLH16 | PGSC0003DMT400040021 | 303 | 34 207.55 | 6.54 | StbHLH54 | PGSC0003DMT400002155 | 312 | 34 997.42 | 6.00 | |
| StbHLH17 | PGSC0003DMT400026937 | 326 | 36 823.76 | 5.23 | StbHLH55 | PGSC0003DMT400033757 | 210 | 23 748.34 | 8.89 | |
| StbHLH18 | PGSC0003DMT400021383 | 244 | 27 458.61 | 5.56 | StbHLH56 | PGSC0003DMT400044959 | 231 | 26 357.79 | 5.64 | |
| StbHLH19 | PGSC0003DMT400026449 | 235 | 26 275.12 | 8.64 | StbHLH57 | PGSC0003DMT400018046 | 578 | 64 285.91 | 7.61 | |
| StbHLH20 | PGSC0003DMT400036021 | 442 | 49 217.42 | 5.90 | StbHLH58 | PGSC0003DMT400040761 | 390 | 44 069.48 | 5.27 | |
| StbHLH21 | PGSC0003DMT400055910 | 267 | 29 914.13 | 7.80 | StbHLH59 | PGSC0003DMT400038134 | 515 | 56 408.13 | 5.13 | |
| StbHLH22 | PGSC0003DMT400078776 | 317 | 35 038.85 | 6.12 | StbHLH60 | PGSC0003DMT400048941 | 312 | 35 241.59 | 4.72 | |
| StbHLH23 | PGSC0003DMT400078675 | 335 | 38 417.26 | 5.44 | StbHLH61 | PGSC0003DMT400040448 | 225 | 25 610.16 | 7.62 | |
| StbHLH24 | PGSC0003DMT400078696 | 347 | 39 529.70 | 6.39 | StbHLH62 | PGSC0003DMT400071861 | 62 | 7 527.78 | 10.4 | |
| StbHLH25 | PGSC0003DMT400078698 | 209 | 23 532.27 | 6.17 | StbHLH63 | PGSC0003DMT400080418 | 260 | 28 511.99 | 7.81 | |
| StbHLH26 | PGSC0003DMT400078699 | 323 | 35 931.15 | 5.47 | StbHLH64 | PGSC0003DMT400012487 | 390 | 42 992.79 | 5.76 | |
| StbHLH27 | PGSC0003DMT400010253 | 376 | 41 285.79 | 5.04 | StbHLH65 | PGSC0003DMT400074075 | 530 | 57 936.63 | 5.31 | |
| StbHLH28 | PGSC0003DMT400034838 | 296 | 32 476.68 | 5.27 | StbHLH66 | PGSC0003DMT400083164 | 416 | 46 807.69 | 8.71 | |
| StbHLH29 | PGSC0003DMT400012997 | 335 | 38 164.49 | 5.23 | StbHLH66 | PGSC0003DMT400069569 | 284 | 31 977.18 | 6.76 | |
| StbHLH30 | PGSC0003DMT400058169 | 268 | 30 772.88 | 7.63 | StbHLH68 | PGSC0003DMT400051980 | 467 | 51 763.96 | 7.19 | |
| StbHLH31 | PGSC0003DMT400048857 | 148 | 16 910.86 | 9.75 | StbHLH69 | PGSC0003DMT400051674 | 477 | 52 903.87 | 6.59 | |
| StbHLH32 | PGSC0003DMT400071293 | 224 | 25 544.12 | 8.60 | StbHLH70 | PGSC0003DMT400028917 | 444 | 46 271.20 | 6.09 | |
| StbHLH33 | PGSC0003DMT400019550 | 214 | 24 405.08 | 8.94 | StbHLH71 | PGSC0003DMT400010463 | 252 | 28 629.96 | 4.97 | |
| StbHLH34 | PGSC0003DMT400023497 | 355 | 38 301.12 | 5.94 | StbHLH72 | PGSC0003DMT400056685 | 374 | 42 308.55 | 8.80 | |
| StbHLH35 | PGSC0003DMT400051137 | 338 | 37 637.08 | 6.08 | StbHLH73 | PGSC0003DMT400041156 | 517 | 57 437.21 | 7.09 | |
| StbHLH36 | PGSC0003DMT400062844 | 291 | 30 828.06 | 5.65 | StbHLH74 | PGSC0003DMT400067287 | 351 | 39 693.02 | 8.14 | |
| StbHLH37 | PGSC0003DMT400063026 | 536 | 58 417.99 | 7.35 | StbHLH75 | PGSC0003DMT400052622 | 255 | 27 880.55 | 6.92 | |
| StbHLH38 | PGSC0003DMT400063117 | 437 | 48 174.88 | 6.79 | StbHLH76 | PGSC0003DMT400018274 | 224 | 25 831.73 | 6.32 | |
| StbHLH77 | PGSC0003DMT400049504 | 485 | 52 745.68 | 6.07 | StbHLH93 | PGSC0003DMT400016793 | 348 | 36 847.65 | 8.57 | |
| StbHLH78 | PGSC0003DMT400002963 | 598 | 65 258.62 | 5.92 | StbHLH94 | PGSC0003DMT400064399 | 388 | 42 575.81 | 5.83 | |
| StbHLH79 | PGSC0003DMT400015017 | 456 | 51 791.64 | 6.22 | StbHLH95 | PGSC0003DMT400028057 | 389 | 44 514.29 | 5.88 | |
| StbHLH80 | PGSC0003DMT400021165 | 435 | 49 379.65 | 5.89 | StbHLH96 | PGSC0003DMT400008370 | 198 | 22 043.97 | 8.88 | |
| StbHLH81 | PGSC0003DMT400053651 | 319 | 36 510.05 | 4.85 | StbHLH97 | PGSC0003DMT400089728 | 305 | 34 320.40 | 5.48 | |
| StbHLH82 | PGSC0003DMT400095303 | 311 | 35 808.26 | 4.90 | StbHLH98 | PGSC0003DMT400076759 | 180 | 20 751.84 | 9.58 | |
| StbHLH83 | PGSC0003DMT400045210 | 330 | 36 732.34 | 6.10 | StbHLH99 | PGSC0003DMT400021130 | 360 | 39 675.21 | 5.92 | |
| StbHLH84 | PGSC0003DMT400045204 | 694 | 75 939.94 | 5.47 | StbHLH100 | PGSC0003DMT400021062 | 380 | 41 565.47 | 9.10 | |
| StbHLH85 | PGSC0003DMT400032139 | 360 | 40 669.07 | 6.81 | StbHLH101 | PGSC0003DMT400041644 | 237 | 27 276.91 | 5.27 | |
| StbHLH86 | PGSC0003DMT400031899 | 431 | 48 790.06 | 6.71 | StbHLH102 | PGSC0003DMT400073760 | 262 | 30 275.48 | 5.87 | |
| StbHLH87 | PGSC0003DMT400053639 | 492 | 53 992.33 | 7.13 | StbHLH103 | PGSC0003DMT400056716 | 441 | 48 896.06 | 6.65 | |
| StbHLH88 | PGSC0003DMT400048756 | 578 | 62 943.67 | 6.42 | StbHLH104 | PGSC0003DMT400020244 | 382 | 39 561.65 | 6.21 | |
| StbHLH89 | PGSC0003DMT400002512 | 284 | 31 495.30 | 5.46 | StbHLH105 | PGSC0003DMT400013766 | 272 | 31 224.18 | 6.76 | |
| StbHLH90 | PGSC0003DMT400033569 | 590 | 64 760.08 | 5.81 | StbHLH106 | PGSC0003DMT400045820 | 296 | 33 899.60 | 7.02 | |
| StbHLH91 | PGSC0003DMT400076223 | 285 | 32 950.42 | 6.27 | StbHLH107 | PGSC0003DMT400011651 | 413 | 46 698.70 | 5.92 | |
| StbHLH92 | PGSC0003DMT400042791 | 439 | 49 242.61 | 6.06 | StbHLH108 | PGSC0003DMT400061284 | 489 | 54 218.28 | 5.12 |

图4 马铃薯bHLH家族基因的染色体定位 1:根;2:叶;3:块茎;4:对照;5:盐;6:甘露醇;7:生长素;8:赤霉素;9:脱落酸;10:热。图中红色代表较对照基因表达上调,蓝色代表较对照基因表达下调
Fig. 4 Chromosome localization of bHLH family genes in S. tuberosum 1:Root. 2:Leaf. 3:Tuber. 4:Control. 5:Salt. 6:Mannitol. 7:Auxin. 8:Gibberellin. 9:Abscisic acid. 10:Heat. In the figure,red refers to the up regulation of gene expression and blue to the down regulation of gene expression
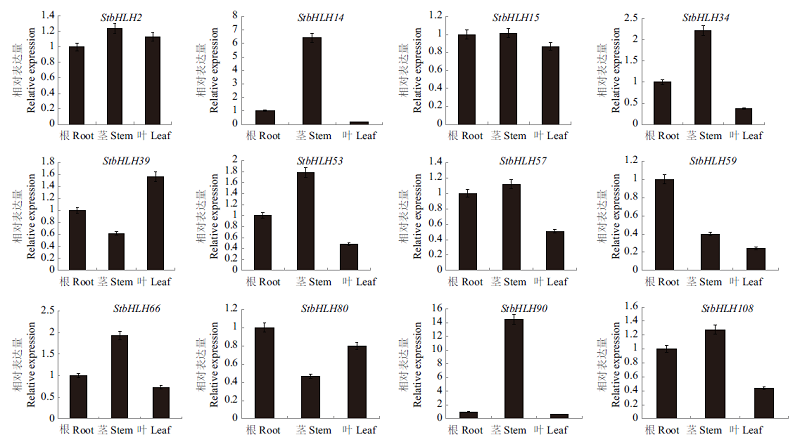
图6 马铃薯bHLH转录因子组织表达模式分析 非生物胁迫包括:低钾、脱落酸、聚乙二醇、高盐、过氧化氢。分别在0、3、6和12 h取样
Fig. 6 Tissue expression pattern analysis of bHLH transcription factor in S. tuberosum Abiotic stresses include low potassium,abscisic acid,polyethylene glycol,high salt and hydrogen peroxide. The samples were taken at 0,3,6 and 12 h, respectively
| [1] |
Niu X, Guan Y, Chen S, et al. Genome-wide analysis of basic helix-loop-helix(bHLH)transcription factors in Brachypodium distachyon[J]. BMC Genomics, 2017, 18(1):619.
doi: 10.1186/s12864-017-4044-4 URL |
| [2] | 张全琪, 朱家红, 倪燕妹, 等. 植物bHLH转录因子的结构特点及其生物学功能[J]. 热带亚热带植物学报, 2011, 19(1):84-90. |
| Zhang QQ, Zhu JH, Ni YM, et al. The structure and function of plant bHLH transcription factors[J]. J Trop Subtrop Bot, 2011, 19(1):84-90. | |
| [3] |
Amoutzias GD, Veron AS, Weiner J, et al. One billion years of bZIP transcription factor evolution:conservation and change in dimerization and DNA-binding site specificity[J]. Mol Biol Evol, 2007, 24(3):827-835.
pmid: 17194801 |
| [4] |
Feller A, Machemer K, Braun EL, et al. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors[J]. Plant J, 2011, 66(1):94-116.
doi: 10.1111/tpj.2011.66.issue-1 URL |
| [5] | 杨鹏程, 周波, 李玉花. 植物花青素合成相关的bHLH转录因子[J]. 植物生理学报, 2012, 48(8):747-758. |
|
Yang PC, Zhou B, Li YH. The bHLH transcription factors involved in anthocyanin biosynjournal in plants[J]. Plant Physiol J, 2012, 48(8):747-758.
doi: 10.1104/pp.48.6.747 URL |
|
| [6] | 陈红霖, 胡亮亮, 王丽侠, 等. 绿豆bHLH转录因子家族的鉴定与生物信息学分析[J]. 植物遗传资源学报, 2017, 18(6):1159-1167. |
| Chen HL, Hu LL, Wang LX, et al. Genome-wide identification and bioinformatics analysis of bHLH transcription factor family in mung bean(Vigna radiata L.)[J]. J Plant Genet Resour, 2017, 18(6):1159-1167. | |
| [7] |
Guo XJ, Wang JR. Global identification, structural analysis and expression characterization of bHLH transcription factors in wheat[J]. BMC Plant Biol, 2017, 17(1):90.
doi: 10.1186/s12870-017-1038-y URL |
| [8] |
Song XM, Huang ZN, Duan WK, et al. Genome-wide analysis of the bHLH transcription factor family in Chinese cabbage(Brassica rapa ssp. pekinensis)[J]. Mol Genet Genom, 2014, 289(1):77-91.
doi: 10.1007/s00438-013-0791-3 URL |
| [9] |
Mao K, Dong Q, Li C, et al. Genome wide identification and characterization of apple bHLH transcription factors and expression analysis in response to drought and salt stress[J]. Front Plant Sci, 2017, 8:480.
doi: 10.3389/fpls.2017.00480 pmid: 28443104 |
| [10] | 惠甜, 沈兵琪, 王连春, 等. 桑树bHLH转录因子家族全基因组鉴定与分析[J]. 分子植物育种, 2019, 17(17):5624-5637. |
| Hui T, Shen BQ, Wang LC, et al. Genome-wide identification and analysis of the bHLH transcription factor gene family in Morus notabilis[J]. Mol Plant Breed, 2019, 17(17):5624-5637. | |
| [11] |
Sun W, Jin X, Ma Z, et al. Basic helix-loop-helix(bHLH)gene family in Tartary buckwheat(Fagopyrum tataricum):Genome-wide identification, phylogeny, evolutionary expansion and expression analyses[J]. Int J Biol Macromol, 2020, 155:1478-1490.
doi: 10.1016/j.ijbiomac.2019.11.126 URL |
| [12] |
Tian S, Li L, Wei M, et al. Genome-wide analysis of basic helix-loop-helix superfamily members related to anthocyanin biosynjournal in eggplant(Solanum melongena L.)[J]. PeerJ, 2019, 7:e7768.
doi: 10.7717/peerj.7768 URL |
| [13] | 何昌文, 朱丽, 沈珊, 等. 银杏bHLH91转录因子基因的克隆及表达分析[J]. 广西植物, 2018, 38(2):202-209. |
| He CW, Zhu L, Shen S, et al. Cloning and expression analysis of a bHLH91 transcription factor gene from Ginkgo biloba[J]. Guihaia, 2018, 38(2):202-209. | |
| [14] |
杨梦婷, 张春, 王作平, 等. 玉米ZmbHLH161基因的克隆及功能研究[J]. 作物学报, 2020, 46(12):2008-2016.
doi: 10.3724/SP.J.1006.2020.03022 |
| Yang MT, Zhang C, Wang ZP, et al. Cloning and functional analysis of ZmbHLH161 gene in maize[J]. Acta Agron Sin, 2020, 46(12):2008-2016. | |
| [15] |
Zhang J, Liu B, Li M, et al. The bHLH transcription factor bHLH104 interacts with IAA-LEUCINE RESISTANT3 and modulates iron homeostasis in Arabidopsis[J]. Plant Cell, 2015, 27(3):787-805.
doi: 10.1105/tpc.114.132704 URL |
| [16] |
Liang G, Zhang H, Li X, et al. bHLH transcription factor bHLH115 regulates iron homeostasis in Arabidopsis thaliana[J]. J Exp Bot, 2017, 68(7):1743-1755.
doi: 10.1093/jxb/erx043 pmid: 28369511 |
| [17] |
Li YY, Sui XY, Yang JS, et al. A novel bHLH transcription factor, NtbHLH1, modulates iron homeostasis in tobacco(Nicotiana tabacum L.)[J]. Biochem Biophys Res Commun, 2020, 522(1):233-239.
doi: 10.1016/j.bbrc.2019.11.063 URL |
| [18] |
Yang XM, Ren YL, Cai Y, et al. Overexpression of OsbHLH107, a member of the basic helix-loop-helix transcription factor family, enhances grain size in rice(Oryza sativa L.)[J]. Rice, 2018, 11(1):41.
doi: 10.1186/s12284-018-0237-y URL |
| [19] |
Zhang L, Kang J, Xie Q, et al. The basic helix-loop-helix transcription factor bHLH95 affects fruit ripening and multiple metabolisms in tomato[J]. J Exp Bot, 2020, 71(20):6311-6327.
doi: 10.1093/jxb/eraa363 URL |
| [20] | 何洁, 顾秀容, 魏春华, 等. 西瓜bHLH转录因子家族基因的鉴定及其在非生物胁迫下的表达分析[J]. 园艺学报, 2016, 43(2):281-294. |
| He J, Gu XR, Wei CH, et al. Identification and expression analysis under abiotic stresses of the bHLH transcription factor gene family in watermelon[J]. Acta Hortic Sin, 2016, 43(2):281-294. | |
| [21] |
Le Hir R, Castelain M, Chakraborti D, et al. AtbHLH68 transcription factor contributes to the regulation of ABA homeostasis and drought stress tolerance in Arabidopsis thaliana[J]. Physiol Plantarum, 2017, 160(3):312-327.
doi: 10.1111/ppl.2017.160.issue-3 URL |
| [22] | Chen HC, Cheng WH, Hong CY, et al. The transcription factor OsbHLH035 mediates seed germination and enables seedling recovery from salt stress through ABA-dependent and ABA-independent pathways, respectively[J]. Rice:N Y, 2018, 11(1):50. |
| [23] | 耿晶晶. 甜橙bHLH家族转录因子发掘及CsbHLH18抗寒功能鉴定与作用机制解析[D]. 武汉:华中农业大学, 2018. |
| Geng JJ. Genome-wide identification of bHLH transcription factor family in sweet orange(Citrus sinensis)and functional characterization and mechanism analysis of CsbHLH18 in cold resistance[D]. Wuhan:Huazhong Agricultural University, 2018. | |
| [24] | 刘玉汇. MYB和bHLH转录因子对马铃薯块茎花色素苷生物合成的调控机理研究[D]. 兰州:甘肃农业大学, 2016. |
| Liu YH. Regulatory mechanism of MYB and bHLH transcription factors on anthocyanin biosynthesis in potato Tuber[D]. Lanzhou:Gansu Agricultural University, 2016. | |
| [25] | Tai HH, Goyer C, Murphy AM. Potato MYB and bHLH transcription factors associated with anthocyanin intensity and common scab resistance[J], 2013, 91(10):722-730. |
| [26] |
Wilkins MR, Gasteiger E, Bairoch A, et al. Protein identification and analysis tools in the ExPASy server[J]. Methods Mol Biol, 1999, 112:531-552.
pmid: 10027275 |
| [27] |
Zhang W, Sun ZR. Random local neighbor joining:a new method for reconstructing phylogenetic trees[J]. Mol Phylogenetics Evol, 2008, 47(1):117-128.
doi: 10.1016/j.ympev.2008.01.019 URL |
| [28] |
Potato Genome Sequencing Consortium, Xu X, Pan SK, et al. Genome sequence and analysis of the Tuber crop potato[J]. Nature, 2011, 475(7355):189-195.
doi: 10.1038/nature10158 URL |
| [29] |
Zhou X, Liao Y, Kim SU, et al. Genome-wide identification and characterization of bHLH family genes from Ginkgo biloba[J]. Sci Rep, 2020, 10(1):13723.
doi: 10.1038/s41598-020-69305-3 URL |
| [30] | 王力伟, 房永雨, 刘红葵, 等. bHLH转录因子的研究进展[J]. 畜牧与饲料科学, 2020, 41(1):23-27. |
| Wang LW, Fang YY, Liu HK, et al. Research progress of bHLH transcription factors[J]. Animal Husb Feed Sci, 2020, 41(1):23-27. | |
| [31] | 何开平, 吴楚. bHLH转录因子对植物形态发生的影响[J]. 安徽农业科学, 2010, 38(35):19957-19959. |
| He KP, Wu C. The effects of bHLH transcription factors on plant morphogenesis[J]. J Anhui Agric Sci, 2010, 38(35):19957-19959. | |
| [32] | 王寻, 陈西霞, 李宏亮, 等. 苹果NLP(Nin-like protein)转录因子基因家族全基因组鉴定及表达模式分析[J]. 中国农业科学, 2019, 52(23):4333-4349. |
| Wang X, Chen XX, Li HL, et al. Genome-wide identification and expression pattern analysis of NLP(Nin-like protein)transcription factor gene family in apple[J]. Sci Agric Sin, 2019, 52(23):4333-4349. | |
| [33] | 陈媞颖, 刘娟, 袁媛, 等. 黄芩bHLH转录因子基因家族生物信息学及表达分析[J]. 中草药, 2018, 49(3):671-677. |
| Chen TY, Liu J, Yuan Y, et al. Analysis of bioinformatics and expression level of bHLH transcription factors in Scutellaria baicalensis[J]. Chin Tradit Herb Drugs, 2018, 49(3):671-677. | |
| [34] | 武明珠, 李锋, 王燃, 等. 烟草转录因子bHLH93基因的克隆及表达分析[J]. 烟草科技, 2015, 48(3):1-7. |
| Wu MZ, Li F, Wang R, et al. Cloning and expression of transcription factor gene b HLH93from Nicotiana tabacum[J]. Tob Sci Technol, 2015, 48(3):1-7. | |
| [35] |
Waseem M, Li N, Su D, et al. Overexpression of a basic helix-loop-helix transcription factor gene, SlbHLH22, promotes early flowering and accelerates fruit ripening in tomato(Solanum lycopersicum L.)[J]. Planta, 2019, 250(1):173-185.
doi: 10.1007/s00425-019-03157-8 URL |
| [1] | 王子颖, 龙晨洁, 范兆宇, 张蕾. 利用酵母双杂交系统筛选水稻中与OsCRK5互作蛋白[J]. 生物技术通报, 2023, 39(9): 117-125. |
| [2] | 刘雯锦, 马瑞, 刘升燕, 杨江伟, 张宁, 司怀军. 马铃薯StCIPK11的克隆及响应干旱胁迫分析[J]. 生物技术通报, 2023, 39(9): 147-155. |
| [3] | 韩浩章, 张丽华, 李素华, 赵荣, 王芳, 王晓立. 盐碱胁迫诱导的猴樟酵母cDNA文库构建及CbP5CS上游调控因子筛选[J]. 生物技术通报, 2023, 39(9): 236-245. |
| [4] | 江润海, 姜冉冉, 朱城强, 侯秀丽. 微生物强化植物修复铅污染土壤的机制研究进展[J]. 生物技术通报, 2023, 39(8): 114-125. |
| [5] | 吕秋谕, 孙培媛, 冉彬, 王佳蕊, 陈庆富, 李洪有. 苦荞转录因子基因FtbHLH3的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 194-203. |
| [6] | 王佳蕊, 孙培媛, 柯瑾, 冉彬, 李洪有. 苦荞糖基转移酶基因FtUGT143的克隆及表达分析[J]. 生物技术通报, 2023, 39(8): 204-212. |
| [7] | 陈晓, 于茗兰, 吴隆坤, 郑晓明, 逄洪波. 植物lncRNA及其对低温胁迫响应的研究进展[J]. 生物技术通报, 2023, 39(7): 1-12. |
| [8] | 王帅, 冯宇梅, 白苗, 杜维俊, 岳爱琴. 大豆GmHMGR基因响应外源激素及非生物胁迫功能研究[J]. 生物技术通报, 2023, 39(7): 131-142. |
| [9] | 孙明慧, 吴琼, 刘丹丹, 焦小雨, 王文杰. 茶树CsTMFs的克隆与表达分析[J]. 生物技术通报, 2023, 39(7): 151-159. |
| [10] | 魏茜雅, 秦中维, 梁腊梅, 林欣琪, 李映志. 褪黑素种子引发处理提高朝天椒耐盐性的作用机制[J]. 生物技术通报, 2023, 39(7): 160-172. |
| [11] | 余慧, 王静, 梁昕昕, 辛亚平, 周军, 赵会君. 宁夏枸杞铁镉响应基因的筛选及其功能验证[J]. 生物技术通报, 2023, 39(7): 195-205. |
| [12] | 张蓓, 任福森, 赵洋, 郭志伟, 孙强, 刘贺娟, 甄俊琦, 王童童, 程相杰. 辣椒响应热胁迫机制的研究进展[J]. 生物技术通报, 2023, 39(7): 37-47. |
| [13] | 丁凯鑫, 王立春, 田国奎, 王海艳, 李凤云, 潘阳, 庞泽, 单莹. 烯效唑缓解植物干旱损伤的研究进展[J]. 生物技术通报, 2023, 39(6): 1-11. |
| [14] | 孔德真, 段震宇, 王刚, 张鑫, 席琳乔. 盐、碱胁迫下高丹草苗期生理特征及转录组学分析[J]. 生物技术通报, 2023, 39(6): 199-207. |
| [15] | 赵雪婷, 高利燕, 王俊刚, 沈庆庆, 张树珍, 李富生. 甘蔗AP2/ERF转录因子基因ShERF3的克隆、表达及其编码蛋白的定位[J]. 生物技术通报, 2023, 39(6): 208-216. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||