








生物技术通报 ›› 2024, Vol. 40 ›› Issue (8): 142-151.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0257
收稿日期:2024-03-16
出版日期:2024-08-26
发布日期:2024-07-31
通讯作者:
屈燕,女,教授,研究方向:园林植物资源利用与开发;E-mail: quyan@swfu.edu.cn作者简介:周麟,男,硕士研究生,研究方向:园林植物资源利用与开发;E-mail: 1617574776@qq.com
基金资助:
ZHOU Lin1( ), HUANG Shun-man1, SU Wen-kun1, YAO Xiang2, QU Yan1(
), HUANG Shun-man1, SU Wen-kun1, YAO Xiang2, QU Yan1( )
)
Received:2024-03-16
Published:2024-08-26
Online:2024-07-31
摘要:
【目的】通过生物信息学方法初步筛选与滇山茶花色相关的bHLH基因,为后续深入研究滇山茶花色提供支持。【方法】基于滇山茶全长转录组数据,通过生物信息学方法鉴定滇山茶bHLH基因家族成员,并对其理化性质、二级结构、系统发育关系和结构域等进行分析。利用二代转录组和代谢组数据挖掘与滇山茶花色相关的bHLH基因,通过实时定量聚合酶链式反应(real-time quantitative polymerase chain reaction, RT-qPCR)验证bHLH基因在滇山茶开花过程中和不同品种间的表达情况。【结果】在滇山茶全长转录组中鉴定到71个滇山茶CrbHLHs基因,蛋白分子量的范围在22 994.58-102 996.96 Da,等电位为4.98-9.51,均为亲水性蛋白。与拟南芥聚类分析发现,滇山茶bHLH基因家族成员可分布到14个亚族。多组学联合分析发现,CrbHLH8、CrbHLH61和CrbHLH63与矢车菊色素苷呈正相关,而CrbHLH59与其呈负相关;CrbHLH13和CrbHLH71与原花青素呈正相关,其中CrbHLH8和CrbHLH61为IIIf亚族成员。【结论】从滇山茶全长转录组中鉴定出71个CrbHLHs家族成员,其中6个CrbHLHs与滇山茶花色相关。
周麟, 黄顺满, 苏文坤, 姚响, 屈燕. 滇山茶bHLH基因家族鉴定及花色形成相关基因筛选[J]. 生物技术通报, 2024, 40(8): 142-151.
ZHOU Lin, HUANG Shun-man, SU Wen-kun, YAO Xiang, QU Yan. Identification of the bHLH Gene Family and Selection of Genes Related to Color Formation in Camellia reticulata[J]. Biotechnology Bulletin, 2024, 40(8): 142-151.
| 基因Gene | 上游引物Forward primer(5'-3') | 下游引物Reverse primer(5'-3') |
|---|---|---|
| CrbHLH8 | GCAGTTGTGCCCTCAACAAG | CACTCACCTCCATTGGCTGT |
| CrbHLH13 | CGGCTGAATGCTCGTTGATG | CTGAAATTGGCGGTGTTGGG |
| CrbHLH59 | ACCCCATTGAATTCCCCACC | TACAAATCCAGGATCCGCGG |
| CrbHLH61 | GCAGTTGTGCCCTCAACAAG | CACTCACCTCCATTGGCTGT |
| CrbHLH63 | TGTCCATTGGATGCTCACCC | CAGCACCATCTTGAGTCCGT |
| CrbHLH71 | AGCCACGTACTGCTGAACAA | TCTGCGCTATTGCCAGAACA |
| CrActin | GTCTGTTCCCTCTCCTCCCT | GAAGGGATCGTTGACAGCGA |
表1 RT-qPCR引物
Table 1 Primers used for RT-qPCR
| 基因Gene | 上游引物Forward primer(5'-3') | 下游引物Reverse primer(5'-3') |
|---|---|---|
| CrbHLH8 | GCAGTTGTGCCCTCAACAAG | CACTCACCTCCATTGGCTGT |
| CrbHLH13 | CGGCTGAATGCTCGTTGATG | CTGAAATTGGCGGTGTTGGG |
| CrbHLH59 | ACCCCATTGAATTCCCCACC | TACAAATCCAGGATCCGCGG |
| CrbHLH61 | GCAGTTGTGCCCTCAACAAG | CACTCACCTCCATTGGCTGT |
| CrbHLH63 | TGTCCATTGGATGCTCACCC | CAGCACCATCTTGAGTCCGT |
| CrbHLH71 | AGCCACGTACTGCTGAACAA | TCTGCGCTATTGCCAGAACA |
| CrActin | GTCTGTTCCCTCTCCTCCCT | GAAGGGATCGTTGACAGCGA |
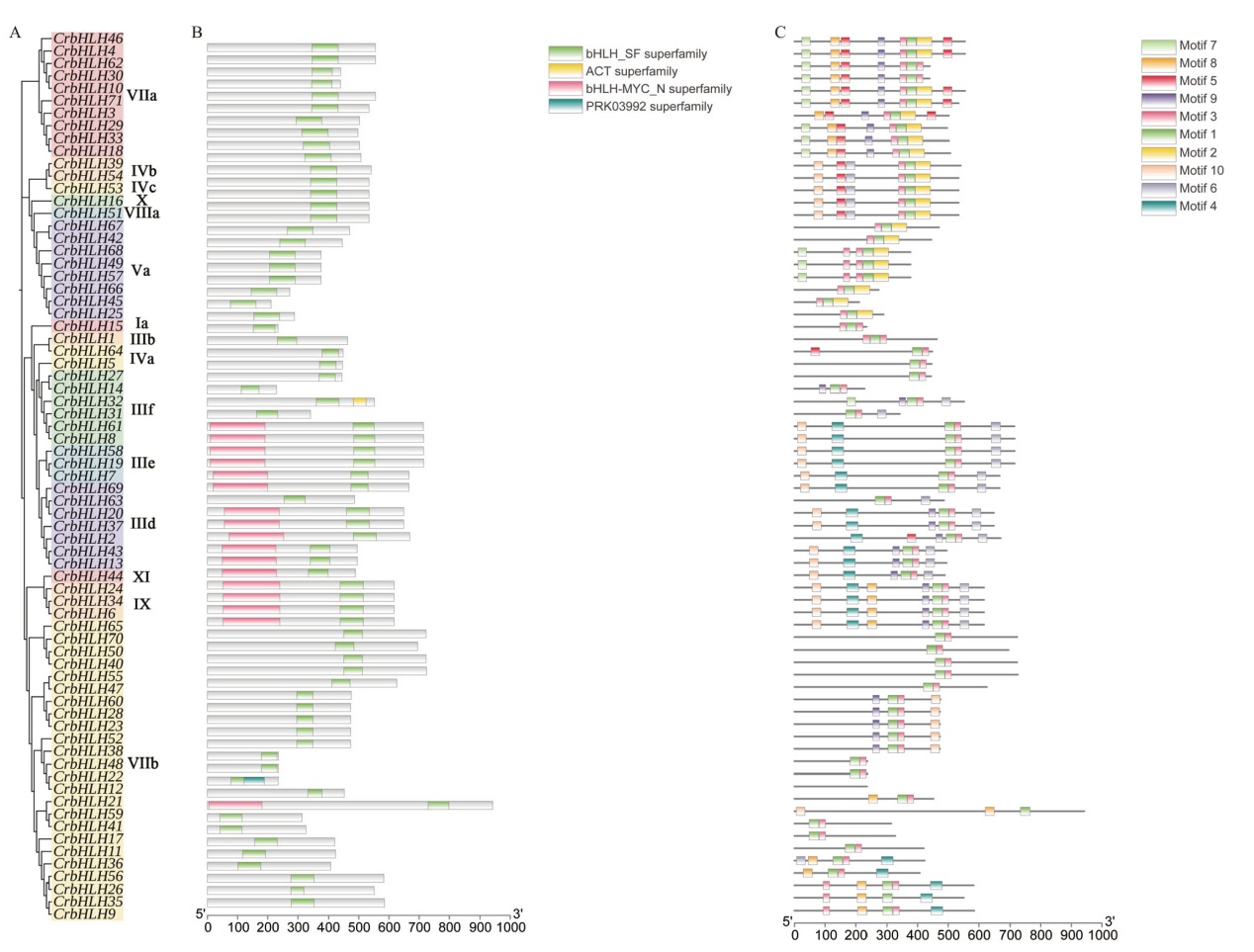
图3 滇山茶bHLH基因家族的保守基序和保守结构域分析 A:滇山茶bHLH基因家族成员聚类分析;B:滇山茶bHLH基因成员的保守基序分析;C:滇山茶bHLH基因家族成员的保守结构域
Fig. 3 Analysis of conserved motif and conserved domain of the bHLH gene family of C. reticulate A: Cluster analysis of bHLH gene family members of C. reticulata. B: Conserved motif analysis of bHLH gene members of C. reticulata. C: Conserved domain of the bHLH gene family members of C. reticulata
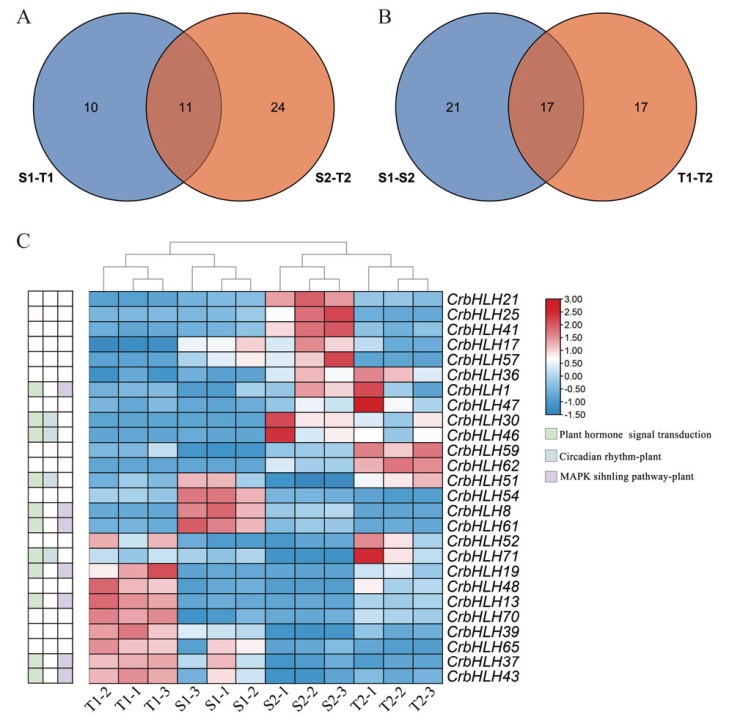
图4 滇山茶差异bHLH分析 A:滇山茶品种间Veen图;B:滇山茶不同发育时期Veen图;C:差异bHLH基因组合热图;S:狮子头;T:童子面;1:花蕾期;2:盛花期;-1、-2、-3表示3个重复。下同
Fig. 4 Differential bHLH analysis of C. reticulate A: Veen map of C. reticulata varieties. B: Veen map of C. reticulata at different developmental stages. C: Differential bHLH gene combination heat map. S: C. reticulata ‘Shizitou’. T: C. reticulata ‘Tongzimian’. 1: Bud stage. 2: Full flowering stage. -1, -2, and -3 indicate three repetitions. The same below
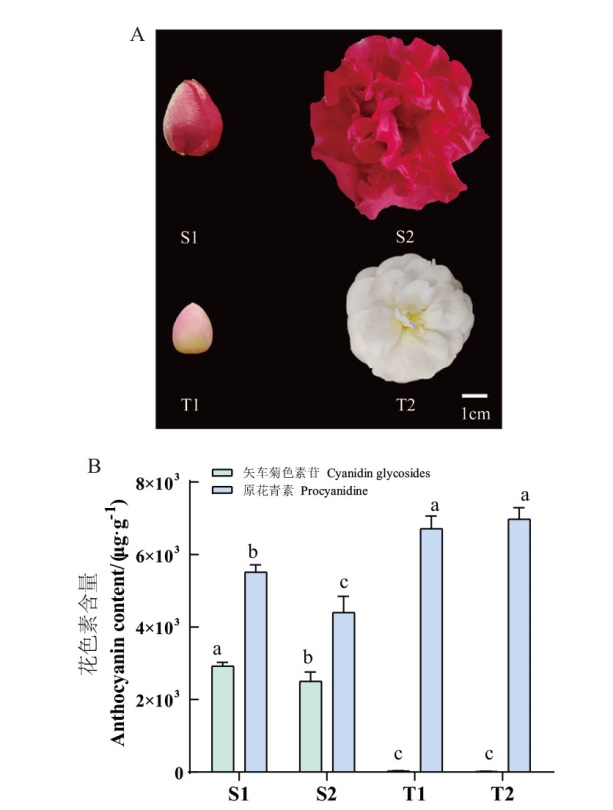
图5 滇山茶‘狮子头’和‘童子面’花色表型(A)及关键色素含量(B) 不同小写字母表示在P<0.05水平差异显著。下同
Fig. 5 Floral phenotype(A), and key pigment content(B)in C. reticulata ‘Shizitou’ and C. reticulata ‘Tongzimian’ Different lower letters indicate significant differences at P<0.05 level. The same below
| [1] | 普梅英, 牛锦华, 付芳丽, 等. 云南山茶枝枯病病原菌分离鉴定[J]. 植物病理学报, 2023, 53(6): 1222-1225. |
| Pu MY, Niu JH, Fu FL, et al. Identification of the pathogen causing dieback disease on Camellia reticulata Lindl. in Yunnan[J]. Acta Phytopathol Sin, 2023, 53(6): 1222-1225. | |
| [2] |
吴贵进, 陈龙清, 韦秋雨, 等. 滇山茶登录品种统计及表型性状分析[J]. 园艺学报, 2023, 50(10): 2157-2170.
doi: 10.16420/j.issn.0513-353x.2022-1192 |
| Wu GJ, Chen LQ, Wei QY, et al. Statistics and phenotypic traits analysis of Camellia reticulata registered cultivars[J]. Acta Hortic Sin, 2023, 50(10): 2157-2170. | |
| [3] | Qu Y, Ou Z, Yong QQ, et al. Coloration differences in three Camellia reticulata Lindl. cultivars: ‘Tongzimian’, ‘Shizitou’ and ‘Damanao’[J]. BMC Plant Biol, 2024, 24(1): 18. |
| [4] | Geng F, Nie RM, Yang N, et al. Integrated transcriptome and metabolome profiling of Camellia reticulata reveal mechanisms of flower color differentiation[J]. Front Genet, 2022, 13: 1059717. |
| [5] |
孙晓波, 郭臻昊, 李畅, 等. 杜鹃花品种大鸳鸯锦花瓣条纹形成的色素基础和差异表达基因的挖掘[J]. 华北农学报, 2023, 38(6): 26-35.
doi: 10.7668/hbnxb.20194067 |
| Sun XB, Guo ZH, Li C, et al. The pigment foundation of the formation of petal stripes in the Rhododendron variety dayuanyangjin and the exploration of differential expression genes[J]. Acta Agric Boreali Sin, 2023, 38(6): 26-35. | |
| [6] | 黄虎, 马先进, 李思佳, 等. 山茶CjMYB1基因的克隆及表达分析[J]. 西北植物学报, 2022, 42(3): 381-389. |
| Huang H, Ma XJ, Li SJ, et al. Molecular cloning and expression analysis of CjMYB1 in Camellia japonica[J]. Acta Bot Boreali Occidentalia Sin, 2022, 42(3): 381-389. | |
| [7] | Li JB, Hashimoto F, Shimizu K, et al. Anthocyanins from red flowers of Camellia reticulata LINDL[J]. Biosci Biotechnol Biochem, 2007, 71(11): 2833-2836. |
| [8] |
安昌, 陆琳, 沈梦千, 等. 植物bHLH基因家族研究进展及在药用植物中的应用前景[J]. 生物技术通报, 2023, 39(10): 1-16.
doi: 10.13560/j.cnki.biotech.bull.1985.2023-0243 |
| An C, Lu L, Shen MQ, et al. Research progress of bHLH gene family in plants and its application prospects in medical plants[J]. Biotechnol Bull, 2023, 39(10): 1-16. | |
| [9] | 杨鹏程, 周波, 李玉花. 植物花青素合成相关的bHLH转录因子[J]. 植物生理学报, 2012, 48(8): 747-758. |
| Yang PC, Zhou B, Li YH. The bHLH transcription factors involved in anthocyanin biosynthesis in plants[J]. Plant Physiol J, 2012, 48(8): 747-758. | |
| [10] | Zhang KK, Lin CY, Chen BY, et al. A light responsive transcription factor CsbHLH89 positively regulates anthocyanidin synthesis in tea(Camellia sinensis)[J]. Sci Hortic, 2024, 327: 112784. |
| [11] |
李博, 刘合霞, 陈宇玲, 等. 金花茶CnbHLH79转录因子的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 241-250.
doi: 10.13560/j.cnki.biotech.bull.1985.2022-1562 |
| Li B, Liu HX, Chen YL, et al. Cloning, subcellular localization and expression analysis of CnbHLH79 transcription factor from Camellia nitidissima[J]. Biotechnol Bull, 2023, 39(8): 241-250. | |
| [12] |
Schaart JG, Dubos C, Romero De La Fuente I, et al. Identification and characterization of MYB-bHLH-WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry(Fragaria × ananassa)fruits[J]. New Phytol, 2013, 197(2): 454-467.
doi: 10.1111/nph.12017 pmid: 23157553 |
| [13] |
Koramutla MK, Ram C, Bhatt D, et al. Genome-wide identification and expression analysis of sucrose synthase genes in allotetraploid Brassica juncea[J]. Gene, 2019, 707: 126-135.
doi: S0378-1119(19)30417-2 pmid: 31026572 |
| [14] | An JP, Zhang XW, Liu YJ, et al. ABI5 regulates ABA-induced anthocyanin biosynthesis by modulating the MYB1-bHLH3 complex in apple[J]. J Exp Bot, 2021, 72(4): 1460-1472. |
| [15] | Shen ZG, Li WY, Li YL, et al. The red flower wintersweet genome provides insights into the evolution of magnoliids and the molecular mechanism for tepal color development[J]. Plant J, 2021, 108(6): 1662-1678. |
| [16] |
Heim MA, Jakoby M, Werber M, et al. The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity[J]. Mol Biol Evol, 2003, 20(5): 735-747.
doi: 10.1093/molbev/msg088 pmid: 12679534 |
| [17] |
葛诗蓓, 张学宁, 韩文炎, 等. 植物类黄酮的生物合成及其抗逆作用机制研究进展[J]. 园艺学报, 2023, 50(1): 209-224.
doi: 10.16420/j.issn.0513-353x.2021-1186 |
|
Ge SB, Zhang XN, Han WY, et al. Research progress on plant flavonoids biosynthesis and their anti-stress mechanism[J]. Acta Hortic Sin, 2023, 50(1): 209-224.
doi: 10.16420/j.issn.0513-353x.2021-1186 |
|
| [18] |
贺威智, 雷伟奇, 郭祥鑫, 等. 百子莲MYB家族鉴定及蓝色形成关键基因功能分析[J]. 园艺学报, 2023, 50(6): 1255-1268.
doi: 10.16420/j.issn.0513-353x.2022-0226 |
| He WZ, Lei WQ, Guo XX, et al. Identification of the MYB gene family and functional analysis of key genes related to blue flower coloration in Agapanthus praecox[J]. Acta Hortic Sin, 2023, 50(6): 1255-1268. | |
| [19] | 龙芬芳. 马缨杜鹃花色苷合成相关MYB基因的筛选与功能初步验证[D]. 贵阳: 贵州大学, 2023: 1-5. |
| Long FF. Screening and preliminary functional verification of MYB gene related to anthocyanin synthesis in Rhododendron camelliottii[D]. Guiyang: Guizhou University, 2023: 1-5. | |
| [20] | 邹红竹, 韩璐璐, 周琳, 等. 滇牡丹MYB家族成员鉴定及PdMYB2的功能验证[J]. 林业科学研究, 2022, 35(5): 1-13. |
| Zou HZ, Han LL, Zhou L, et al. Identification of MYB family members and functional verification of PdMYB2 in Paeonia delavayi[J]. For Res, 2022, 35(5): 1-13. | |
| [21] |
范文广, 李保豫, 田辉, 等. 兰州百合鳞茎花青素合成相关MYB与bHLH转录因子的筛选分析[J]. 食品与发酵工业, 2024, 50(2): 57-66.
doi: 10.13995/j.cnki.11-1802/ts.033270 |
| Fan WG, Li BY, Tian H, et al. Screening and analysis of MYB and bHLH transcription factors associated with anthocyanin synthesis in Lilium davidii var. unicolor bulbs[J]. Food Ferment Ind, 2024, 50(2): 57-66. | |
| [22] | 王洪飞, 欧静, 王孝敬, 等. 马缨杜鹃bHLH转录因子家族的鉴定与表达分析[J/OL]. 广西植物, 2023. https://kns.cnki.net/kcms/detail/45.1134.Q.20230720.2036.002.html. |
| Wang HF, Ou J, Wang XJ, et al. Identification and expression analysis of bHLH transcription factors family in Rhododendron delavayi[J/OL]. Guihaia, 2023. https://kns.cnki.net/kcms/detail/45.1134.Q.20230720.2036.002.html. | |
| [23] | 陈丽飞, 刘云怡慧, 李嘉峻, 等. 干旱胁迫下大苞萱草bHLH转录因子家族鉴定与分析[J]. 西南农业学报, 2023, 36(5): 1027-1038. |
| Chen LF, Liu Y, Li JJ, et al. Identification and analysis of bHLH transcription factor gene family in Hemerocallis middendorffii under drought stress[J]. Southwest China J Agric Sci, 2023, 36(5): 1027-1038. | |
| [24] | Li XJ, Cao LJ, Jiao BB, et al. The bHLH transcription factor AcB2 regulates anthocyanin biosynthesis in onion(Allium cepa L.)[J]. Hortic Res, 2022, 9: uhac128. |
| [25] | 陈佳红, 蒋玲莉, 钱婕妤, 等. 百日草bHLH家族IIIf亚族基因筛选及表达分析[J]. 农业生物技术学报, 2023, 31(1): 61-72. |
| Chen JH, Jiang LL, Qian JY, et al. Screening and expression analysis of IIIf subgroup gene of bHLH family in Zinnia elegans[J]. J Agric Biotechnol, 2023, 31(1): 61-72. | |
| [26] | Song QL, Gong WF, Yu XR, et al. Transcriptome and anatomical comparisons reveal the effects of methyl jasmonate on the seed development of Camellia oleifera[J]. J Agric Food Chem, 2023, 71(17): 6747-6762. |
| [27] |
Dombrecht B, Xue GP, Sprague SJ, et al. MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis[J]. Plant Cell, 2007, 19(7): 2225-2245.
doi: 10.1105/tpc.106.048017 pmid: 17616737 |
| [28] |
Lorenzo O, Chico JM, Sánchez-Serrano JJ, et al. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis[J]. Plant Cell, 2004, 16(7): 1938-1950.
doi: 10.1105/tpc.022319 pmid: 15208388 |
| [29] | Schweizer F, Fernández-Calvo P, Zander M, et al. Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC 4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior[J]. Plant Cell, 2013, 25(8): 3117-3132. |
| [30] | 周麟, 雍清青, 姚响, 等. 滇山茶狮子头及其芽变品种大玛瑙间花色差异分析[J]. 植物遗传资源学报, 2024, 25(4): 960-971. |
| Zhou L, Yong QQ, Yao X, et al. Analysis of flower color difference between Camellia reticulata ‘shizitou’ and its bud mutant ‘damanao’[J]. J Plant Genet Resour, 2024, 25(4): 960-971. | |
| [31] | Qi Y, Zhou L, Han LL, et al. PsbHLH1, a novel transcription factor involved in regulating anthocyanin biosynthesis in tree peony(Paeonia suffruticosa)[J]. Plant Physiol Biochem, 2020, 154: 396-408. |
| [32] |
Tao RY, Yu WJ, Gao YH, et al. Light-induced basic/helix-loop-helix64 enhances anthocyanin biosynthesis and undergoes CONSTITUTIVELY PHOTOMORPHOGENIC1-mediated degradation in pear[J]. Plant Physiol, 2020, 184(4): 1684-1701.
doi: 10.1104/pp.20.01188 pmid: 33093233 |
| [33] |
吕秋谕, 孙培媛, 冉彬, 等. 苦荞转录因子基因FtbHLH3的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 194-203.
doi: 10.13560/j.cnki.biotech.bull.1985.2023-0034 |
| Lyu QY, Sun PY, Ran B, et al. Cloning, subcellular localization and expression analysis of the transcription factor gene FtbHLH3 in Fagopyrum tataricum[J]. Biotechnol Bull, 2023, 39(8): 194-203. |
| [1] | 李雨晴, 吴楠, 罗建让. 卵叶牡丹花色苷合成相关基因bHLH的克隆与功能分析[J]. 生物技术通报, 2024, 40(8): 174-185. |
| [2] | 聂祝欣, 郭瑾, 乔子洋, 李微薇, 张学燕, 刘春阳, 王静. 黑果枸杞不同发育时期果实花色苷合成的转录组分析[J]. 生物技术通报, 2024, 40(8): 106-117. |
| [3] | 杨巍, 赵丽芬, 唐兵, 周麟笔, 杨娟, 莫传园, 张宝会, 李飞, 阮松林, 邓英. 芥菜SRO基因家族全基因组鉴定与表达分析[J]. 生物技术通报, 2024, 40(8): 129-141. |
| [4] | 武帅, 辛燕妮, 买春海, 穆晓娅, 王敏, 岳爱琴, 赵晋忠, 吴慎杰, 杜维俊, 王利祥. 大豆GS基因家族全基因组鉴定及胁迫响应分析[J]. 生物技术通报, 2024, 40(8): 63-73. |
| [5] | 张明亚, 庞胜群, 刘玉东, 苏永峰, 牛博文, 韩琼琼. 番茄FAD基因家族的鉴定与表达分析[J]. 生物技术通报, 2024, 40(7): 150-162. |
| [6] | 臧文蕊, 马明, 砗根, 哈斯阿古拉. 甜瓜BZR转录因子家族基因的全基因组鉴定及表达模式分析[J]. 生物技术通报, 2024, 40(7): 163-171. |
| [7] | 王健, 杨莎, 孙庆文, 陈宏宇, 杨涛, 黄园. 金钗石斛bHLH转录因子家族全基因组鉴定及表达分析[J]. 生物技术通报, 2024, 40(6): 203-218. |
| [8] | 李梦然, 叶伟, 李赛妮, 张维阳, 李建军, 章卫民. Lithocarols类化合物生物合成基因litI的表达及其启动子功能分析[J]. 生物技术通报, 2024, 40(6): 310-318. |
| [9] | 胡永波, 雷雨田, 杨永森, 陈馨, 林黄昉, 林碧英, 刘爽, 毕格, 申宝营. 黄瓜和南瓜Bcl-2相关抗凋亡家族全基因组鉴定与表达模式分析[J]. 生物技术通报, 2024, 40(6): 219-237. |
| [10] | 常雪瑞, 王田田, 王静. 辣椒E2基因家族的鉴定及分析[J]. 生物技术通报, 2024, 40(6): 238-250. |
| [11] | 刘蓉, 田闵玉, 李光泽, 谭成方, 阮颖, 刘春林. 甘蓝型油菜REVEILLE家族鉴定及诱导表达分析[J]. 生物技术通报, 2024, 40(6): 161-171. |
| [12] | 侯雅琼, 郎红珊, 闻蒙蒙, 谷易云, 朱润洁, 汤晓丽. 猕猴桃AcHSP20基因家族的鉴定及表达分析[J]. 生物技术通报, 2024, 40(5): 167-178. |
| [13] | 李嘉欣, 李鸿燕, 刘丽娥, 张恬, 周武. 沙棘NRAMP基因家族鉴定及铅胁迫下表达分析[J]. 生物技术通报, 2024, 40(5): 191-202. |
| [14] | 肖雅茹, 贾婷婷, 罗丹, 武喆, 李丽霞. 黄瓜CsERF025L转录因子的克隆及表达分析[J]. 生物技术通报, 2024, 40(4): 159-166. |
| [15] | 钟匀, 林春, 刘正杰, 董陈文华, 毛自朝, 李兴玉. 芦笋皂苷合成相关糖基转移酶基因克隆及原核表达分析[J]. 生物技术通报, 2024, 40(4): 255-263. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||