








生物技术通报 ›› 2025, Vol. 41 ›› Issue (10): 186-195.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0421
翁慧婷1,2( ), 郭惠明3, 程红梅3, 李君2, 张超2, 刘海洋1(
), 郭惠明3, 程红梅3, 李君2, 张超2, 刘海洋1( ), 苏晓峰3(
), 苏晓峰3( )
)
收稿日期:2025-04-22
出版日期:2025-10-26
发布日期:2025-10-28
通讯作者:
苏晓峰,男,博士,研究员,研究方向 :植物与微生物互作;E-mail: suxiaofeng@caas.cn;作者简介:翁慧婷,女,博士研究生,研究方向 :植物学;E-mail: wht_bio@163.com
基金资助:
WENG Hui-ting1,2( ), GUO Hui-ming3, CHENG Hong-mei3, LI Jun2, ZHANG Chao2, LIU Hai-yang1(
), GUO Hui-ming3, CHENG Hong-mei3, LI Jun2, ZHANG Chao2, LIU Hai-yang1( ), SU Xiao-feng3(
), SU Xiao-feng3( )
)
Received:2025-04-22
Published:2025-10-26
Online:2025-10-28
摘要:
目的 建立一种同时检测4种黄萎病致病菌的微滴式数字PCR(ddPCR)方法,为及时、准确定量监测该病原真菌的生长动态,进行早期诊断和风险评估奠定基础。 方法 通过比对4种黄萎病致病菌大丽轮枝菌(Verticillium dahliae, Vd)、长孢轮枝菌(V. longisporum, Vl)、非苜蓿轮枝菌(V. nonalfalfae, Vna)和黑白轮枝菌(V. albo-atrum, Vaa)的ITS(Internal transcribed spacer)序列(Vd,KY039312.1;Vl,KX058040.1;Vna,KT362917.1和Vaa,MH856937.1),选取保守区域设计引物和探针。结合微滴式数字PCR和实时荧光定量PCR(qPCR)筛选最佳引物,优化ddPCR最佳反应体系,并测定方法的特异性与灵敏度。 结果 建立方法的最佳引物/探针组为Ver5;最佳退火温度为58 ℃,引物浓度为500 nmol/L和探针浓度为250 nmol/L。特异性检测结果显示,该方法能够特异性识别4种黄萎病致病菌,对包括7种真菌和6种细菌在内的非靶标微生物无交叉扩增;对于Vd、Vl、Vna、Vaa的检测限分别为2.1×10-6、1.6×10-6、6.9×10-4、3.6×10-5 ng/μL。选取50个棉花和50份土壤样品展开检测分析,结果表明相较于qPCR,ddPCR方法的检出率呈现出显著优势,且检测灵敏度分别提高了46%和51%。 结论 建立的ddPCR方法检测4种黄萎病致病菌特异性强,灵敏度高,稳定可靠,为黄萎病的精准检测提供了重要的技术手段。该方法有利于海关检验检疫与植物病虫害监管等领域,提高病害防控的科学性与时效性。
翁慧婷, 郭惠明, 程红梅, 李君, 张超, 刘海洋, 苏晓峰. 四种主要黄萎病致病菌微滴数字PCR检测方法的建立及应用[J]. 生物技术通报, 2025, 41(10): 186-195.
WENG Hui-ting, GUO Hui-ming, CHENG Hong-mei, LI Jun, ZHANG Chao, LIU Hai-yang, SU Xiao-feng. Establishment and Application of Droplet Digital PCR Detection Methods for Four Major Verticillium Wilt Pathogens[J]. Biotechnology Bulletin, 2025, 41(10): 186-195.
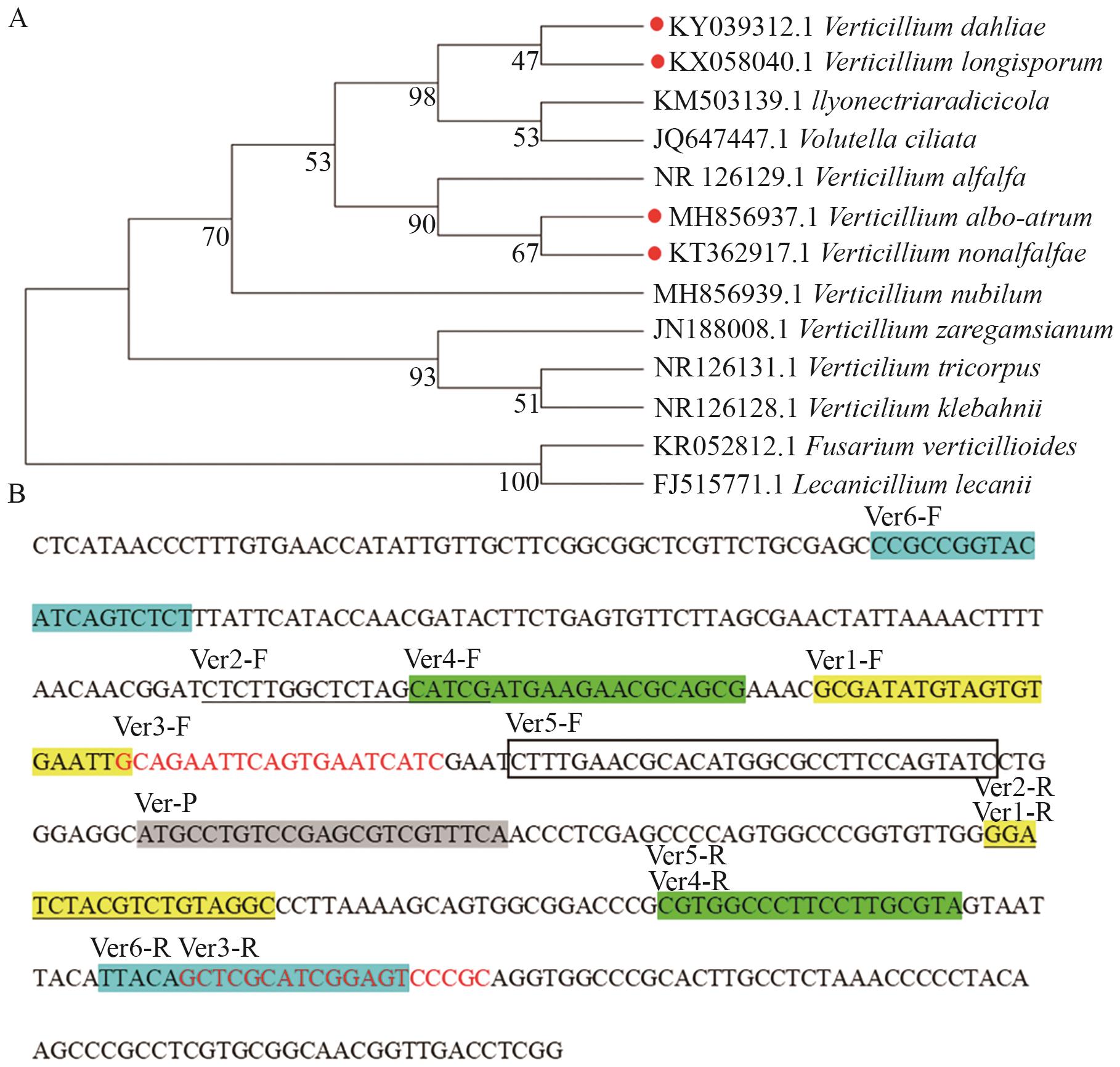
图1 进化树分析及引物和探针在ITS序列中的位置A:轮枝菌属及其他真菌的进化树构建; B:引物和探针在ITS序列中的位置
Fig. 1 Phylogenetic analysis and the position of primers and probes in ITS sequencesA: Construction of the phylogenetic tree for Verticillium and other fungi; B: position of primers and probes in ITS sequence
引物名称 Primer name | 序列 Sequence (5′-3′) | 长度 Length (bp) |
|---|---|---|
| Ver1 | F:GCGATATGTAGTGTGAATTG | 152 |
| R:GCCTACAGACGTAGATCC | ||
| P:ATGCCTGTCCGAGCGTCGTTTCA | ||
| Ver2 | F:CTCTTGGCTCTAGCATCG | 189 |
| R:GCCTACAGACGTAGATCC | ||
| P:ATGCCTGTCCGAGCGTCGTTTCA | ||
| Ver3 | F:GCAGAATTCAGTGAATCATC | 203 |
| R:GCGGGACTCCGATGCGAGC | ||
| P:ATGCCTGTCCGAGCGTCGTTTCA | ||
| Ver4 | F:CATCGATGAAGAACGCAGCG | 97 |
| R:TACGCAAGGAAGGGCCACG | ||
| P:ATGCCTGTCCGAGCGTCGTTTCA | ||
| Ver5 | F:CATCGATGAAGAACGCAGCG | 218 |
| R:TACGCAAGGAAGGGCCACG | ||
| P:ATGCCTGTCCGAGCGTCGTTTCA | ||
| Ver6 | F:CCGCCGGTACATCAGTCTCT | 337 |
| R:ACTCCGATGCGAGCTGTAA | ||
| P:ATGCCTGTCCGAGCGTCGTTTCA |
表1 本研究所使用的引物和探针
Table 1 Primers and probes used in this study
引物名称 Primer name | 序列 Sequence (5′-3′) | 长度 Length (bp) |
|---|---|---|
| Ver1 | F:GCGATATGTAGTGTGAATTG | 152 |
| R:GCCTACAGACGTAGATCC | ||
| P:ATGCCTGTCCGAGCGTCGTTTCA | ||
| Ver2 | F:CTCTTGGCTCTAGCATCG | 189 |
| R:GCCTACAGACGTAGATCC | ||
| P:ATGCCTGTCCGAGCGTCGTTTCA | ||
| Ver3 | F:GCAGAATTCAGTGAATCATC | 203 |
| R:GCGGGACTCCGATGCGAGC | ||
| P:ATGCCTGTCCGAGCGTCGTTTCA | ||
| Ver4 | F:CATCGATGAAGAACGCAGCG | 97 |
| R:TACGCAAGGAAGGGCCACG | ||
| P:ATGCCTGTCCGAGCGTCGTTTCA | ||
| Ver5 | F:CATCGATGAAGAACGCAGCG | 218 |
| R:TACGCAAGGAAGGGCCACG | ||
| P:ATGCCTGTCCGAGCGTCGTTTCA | ||
| Ver6 | F:CCGCCGGTACATCAGTCTCT | 337 |
| R:ACTCCGATGCGAGCTGTAA | ||
| P:ATGCCTGTCCGAGCGTCGTTTCA |

图2 琼脂糖凝胶电泳分析M:DNA marker DL5 000;1-4:分别为Vd、Vl、Vna、Vaa基因组DNA
Fig. 2 Analysis via agarose gel electrophoresisM: DNA marker DL5 000; 1-4: indicate the genomic DNA of Vd, Vl, Vna, and Vaa, respectively
菌株DNA Strain DNA | DNA浓度 DNA concentration (ng/μL) | Ver1 | Ver2 | Ver3 | Ver4 | Ver5 | Ver6 |
|---|---|---|---|---|---|---|---|
| Vd | 18.2 | 19.67 | 17.70 | 18.28 | 18.63 | 15.02 | 16.24 |
| Vl | 19 | 15.65 | 13.53 | 14.42 | 14.53 | 11.19 | 11.93 |
| Vna | 22.2 | 17.96 | 16.04 | 16.70 | 16.99 | 13.48 | 14.25 |
| Vaa | 23.8 | 17.19 | 15.14 | 15.85 | 16.13 | 12.49 | 14.07 |
阴性对照 Negative control | 0 | Un. | 37 | 37 | Un. | 37 | 37 |
表2 不同引物和探针的Ct值
Table 2 Ct values in different sets of primers and probes
菌株DNA Strain DNA | DNA浓度 DNA concentration (ng/μL) | Ver1 | Ver2 | Ver3 | Ver4 | Ver5 | Ver6 |
|---|---|---|---|---|---|---|---|
| Vd | 18.2 | 19.67 | 17.70 | 18.28 | 18.63 | 15.02 | 16.24 |
| Vl | 19 | 15.65 | 13.53 | 14.42 | 14.53 | 11.19 | 11.93 |
| Vna | 22.2 | 17.96 | 16.04 | 16.70 | 16.99 | 13.48 | 14.25 |
| Vaa | 23.8 | 17.19 | 15.14 | 15.85 | 16.13 | 12.49 | 14.07 |
阴性对照 Negative control | 0 | Un. | 37 | 37 | Un. | 37 | 37 |
植物病原体 Plant pathogen | Ct值 Ct value | ||||
|---|---|---|---|---|---|
| Ver2 | Ver3 | Ver5 | Ver6 | ||
稻瘟病菌 Magnaporthe oryzae | Un. | Un. | Un. | Un. | |
玉米小斑病菌 Bipolaris maydis | Un. | Un. | Un. | Un. | |
玉米大斑病菌 Exserohilum turcicum | Un. | Un. | Un. | Un. | |
尖孢镰刀菌粘团专化型 Fusarium fujikuroi f. sp. nirenbergiae | Un. | Un. | Un. | Un. | |
禾谷丝核菌 Rhizoctonia solani | Un. | Un. | Un. | Un. | |
南方根结线虫 Meloidogyne incognita | Un. | Un. | Un. | Un. | |
假禾谷镰孢菌 Fusarium pseudograminearum | Un. | Un. | Un. | Un. | |
水稻稻曲病菌 Ustilaginoidea virens | Un. | Un. | Un. | Un. | |
西瓜嗜酸菌 Acidovorax citrulli | Un. | Un. | Un. | Un. | |
水稻白叶枯病菌 Xanthomonas oryzae pv. oryzae | Un. | Un. | Un. | Un. | |
丁香假单胞菌 Pseudomonas syringae | Un. | Un. | Un. | Un. | |
青枯菌 Ralstonia solanacearum | Un. | Un. | Un. | Un. | |
野油菜黄单胞菌 Xanthomonas campestris pv. campestris | Un. | Un. | Un. | Un. | |
表3 引物/探针特异性分析
Table 3 Specific analysis of primers/probe
植物病原体 Plant pathogen | Ct值 Ct value | ||||
|---|---|---|---|---|---|
| Ver2 | Ver3 | Ver5 | Ver6 | ||
稻瘟病菌 Magnaporthe oryzae | Un. | Un. | Un. | Un. | |
玉米小斑病菌 Bipolaris maydis | Un. | Un. | Un. | Un. | |
玉米大斑病菌 Exserohilum turcicum | Un. | Un. | Un. | Un. | |
尖孢镰刀菌粘团专化型 Fusarium fujikuroi f. sp. nirenbergiae | Un. | Un. | Un. | Un. | |
禾谷丝核菌 Rhizoctonia solani | Un. | Un. | Un. | Un. | |
南方根结线虫 Meloidogyne incognita | Un. | Un. | Un. | Un. | |
假禾谷镰孢菌 Fusarium pseudograminearum | Un. | Un. | Un. | Un. | |
水稻稻曲病菌 Ustilaginoidea virens | Un. | Un. | Un. | Un. | |
西瓜嗜酸菌 Acidovorax citrulli | Un. | Un. | Un. | Un. | |
水稻白叶枯病菌 Xanthomonas oryzae pv. oryzae | Un. | Un. | Un. | Un. | |
丁香假单胞菌 Pseudomonas syringae | Un. | Un. | Un. | Un. | |
青枯菌 Ralstonia solanacearum | Un. | Un. | Un. | Un. | |
野油菜黄单胞菌 Xanthomonas campestris pv. campestris | Un. | Un. | Un. | Un. | |
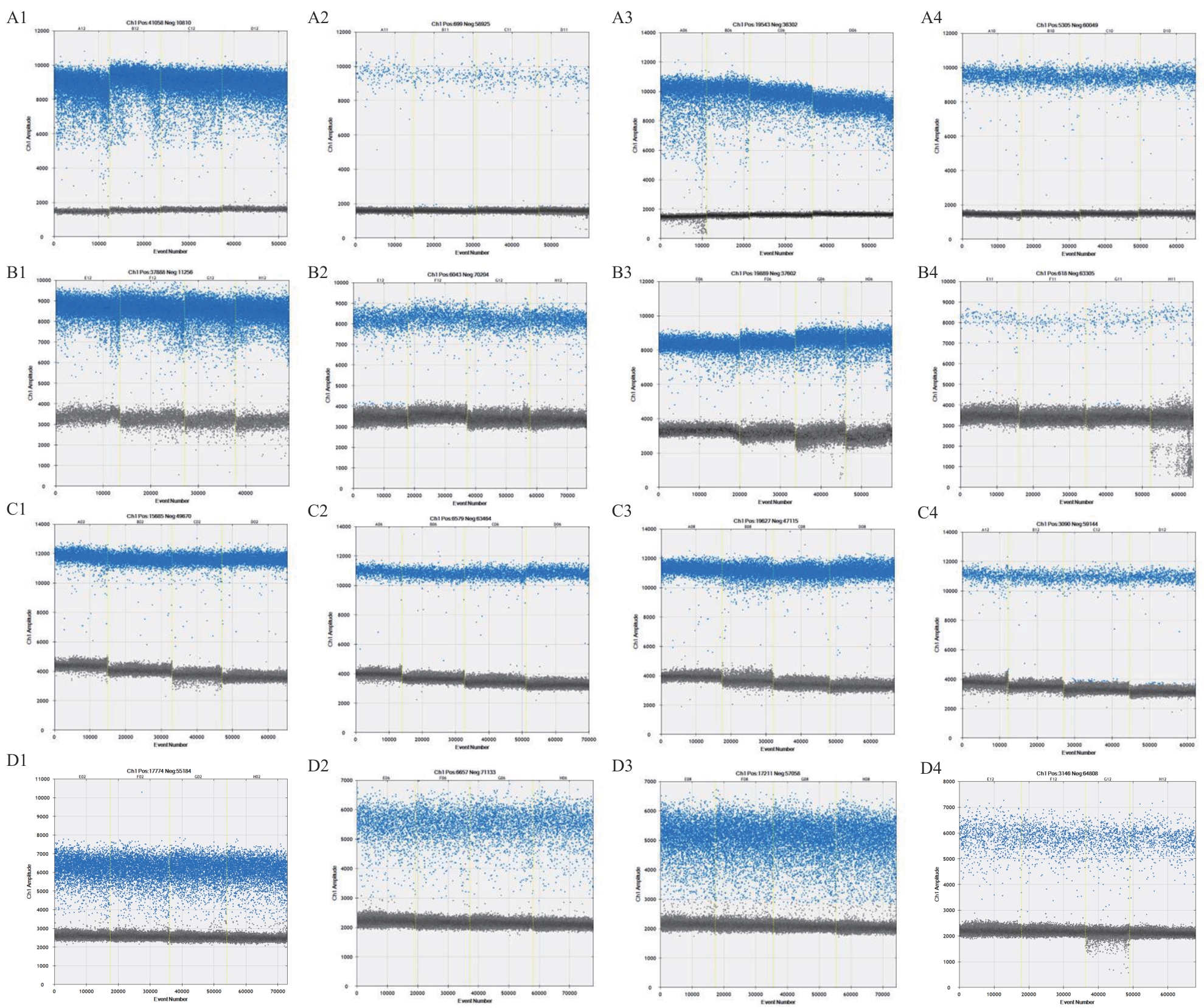
图3 不同引物和探针的ddPCR检测结果A-D:分别为Ver2、Ver3、Ver5和Ver6进行ddPCR试验的引物组合展示。A1-D1:Vd液滴图;A2-D2:Vl液滴图;A3-D3:Vna液滴图,A4-D4:Vaa液滴图
Fig. 3 ddPCR plots in different sets of primers and probesA-D: Primer combinations of ddPCR tests for Ver2, Ver3, Ver5 and Ver6, respectively. A1-D1: The raindrop map of Vd; A2-D2: the raindrop map of Vl; A3-D3: the raindrop pattern of Vna; A4-D4: the raindrop pattern of Vaa

图4 Vd(A)、Vl(B)、Vna(C)、Vaa(D)不同退火温度下的ddPCR结果10个ddPCR反应用黄色纵虚线划分,退火温度梯度为54-62 ℃。蓝色是阳性微滴,灰色是阴性微滴
Fig. 4 ddPCR results of Vd (A), Vl (B), Vna (C) and Vaa (D) at different annealing temperaturesThe 10 ddPCR reactions are divided by yellow dotted lines, and the annealing temperature gradient is 54-62 ℃. The blue is the positive drop, and gray is the negative drop

图5 Vd(A)、Vl(B)、Vna(C)和Vaa(D)不同浓度引物和探针的ddPCR结果6个ddPCR反应用垂直黄色虚线划分,引物梯度范围为400-600 nmol/L,探针梯度范围为100-400 nmol/L。蓝色为阳性微滴,灰色为阴性微滴
Fig. 5 ddPCR results with different concentration of primers and probe in Vd (A), Vl (B), Vna (C), and Vaa (D)Six ddPCR reactions are divided by vertical dotted yellow lines with a primer gradient ranged from 400-600 nmol/L, and a probe gradient ranged from100-400 nmol/L. The blue is the positive drop, and gray is the negative drop

图6 ddPCR的特异性检测1:Vd;2:Vl;3:Vna;4:Vaa;5:M. oryzae;6:B. maydis;7:E. turcicum;8:Foc;9:R. cerealis;10:M. incognita;11:F. pseudograminearum;12:U. virens;13:A. citrulli;14:Xoo;15:P. syringae;16:R. solanacearum;17:Xcc
Fig. 6 Specific detection of ddPCR

图7 ddPCR灵敏度检测结果以Vd(A)、Vl(B)、Vna(C)和Vaa(D)基因组DNA为模板。76个ddPCR反应用黄虚线垂直划分,菌液浓度分别为2.1 × 10-3 ng/μL-2.1 × 10-7 ng/μL、1.6 × 10-3 ng/μL-1.6 × 10-6 ng/μL、6.9 × 10-2 ng/μL-6.9 × 10-6 ng/μL和3.6 × 10-3 ng/μL-3.6 × 10-7 ng/μL。粉红色的线是阈值,蓝色部分为阳性液滴,灰色部分为阴性液滴
Fig. 7 Test results of ddPCR sensitivityThe Vd (A), Vl (B), Vna (C) and Vaa (D) genomic DNAs were used as template. Seventy-six ddPCR reactions are divided by vertical dotted yellow lines with bacteria solution concentration from 2.1 × 10-3 ng/μL to 2.1 × 10-7 ng/μL, 1.6 × 10-3 ng/μL to 1.6 × 10-6 ng/μL, 6.9 × 10-2 ng/μL to 6.9 × 10-6 ng/μL and 3.6 × 10-3 ng/μL to 3.6 × 10-7 ng/μL, respectively. The pink line is the threshold, blue part is positive droplet and gray part is negative droplet
样本 Sample | 分析 Analysis | 阳性/全部(阳性率%) Positive/total(% Positive) |
|---|---|---|
植物 Plant | qPCR | 15/50(30%) |
| ddPCR | 22/50(44%) | |
土壤 Soil | qPCR | 27/50(54%) |
| ddPCR | 41/50(82%) |
表4 qPCR和ddPCR检测50份棉花和土壤样本
Table 4 Detection of 50 cotton and soil samples using qPCR and ddPCR
样本 Sample | 分析 Analysis | 阳性/全部(阳性率%) Positive/total(% Positive) |
|---|---|---|
植物 Plant | qPCR | 15/50(30%) |
| ddPCR | 22/50(44%) | |
土壤 Soil | qPCR | 27/50(54%) |
| ddPCR | 41/50(82%) |
| [1] | Klosterman SJ, Subbarao KV, Kang S, et al. Comparative genomics yields insights into niche adaptation of plant vascular wilt pathogens [J]. PLoS Pathog, 2011, 7(7): e1002137. |
| [2] | Pegg G F, Brady B L. Verticillium wilts [M]. New York: CABI Publishing, 2002. |
| [3] | Depotter JRL, Deketelaere S, Inderbitzin P, et al. Verticillium longisporum, the invisible threat to oilseed rape and other brassicaceous plant hosts [J]. Mol Plant Pathol, 2016, 17(7): 1004-1016. |
| [4] | Flajšman M, Radišek S, Javornik B. Pathogenicity assay of Verticillium nonalfalfae on hop plants [J]. Bio Protoc, 2017, 7(6): e2171. |
| [5] | Larsen RC, Vandemark GJ, Hughes TJ, et al. Development of a real-time polymerase chain reaction assay for quantifying Verticillium albo-atrum DNA in resistant and susceptible alfalfa [J]. Phytopathology, 2007, 97(11): 1519-1525. |
| [6] | Duressa D, Rauscher G, Koike ST, et al. A real-time PCR assay for detection and quantification of Verticillium dahliae in spinach seed [J]. Phytopathology, 2012, 102(4): 443-451. |
| [7] | Floerl S, Druebert C, Majcherczyk A, et al. Defence reactions in the apoplastic proteome of oilseed rape (Brassica napus var. napus) attenuate Verticillium longisporum growth but not disease symptoms [J]. BMC Plant Biol, 2008, 8: 129. |
| [8] | Wang D, Chen JY, Song J, et al. Cytotoxic function of xylanase VdXyn4 in the plant vascular wilt pathogen Verticillium dahliae [J]. Plant Physiol, 2021, 187(1): 409-429. |
| [9] | 索南措, 黄远志, 李彦忠, 等. 苜蓿黄萎病的发生、危害及检测 [J]. 草业科学, 2019, 36(9): 2384-2394. |
| Suo NC, Huang YZ, Li YZ, et al. Verticillium wilt of alfalfa: Occurrence, perniciousness, and detection[J]. Pratacultural Science, 2019, 36(9): 2384-2394 . | |
| [10] | Radišek S, Jakše J, Javornik B. Development of pathotype-specific SCAR markers for detection of Verticillium albo-atrum isolates from hop [J]. Plant Dis, 2004, 88(10): 1115-1122. |
| [11] | Griffiths DA. The development of lignitubers in roots after infection by Verticillium dahliae Kleb [J]. Can J Microbiol, 1971, 17(4): 441-444. |
| [12] | Isaac I. Verticillium wilt of sainfoin [J]. Ann Appl Biol, 1946, 33(1): 28-34. |
| [13] | Mercado-Blanco J, Rodríguez-Jurado D, Pérez-Artés E, et al. Detection of the nondefoliating pathotype of Verticillium dahliae in infected olive plants by nested PCR [J]. Plant Pathol, 2001, 50(5): 609-619. |
| [14] | Huang HC. Verticillium wilt of alfalfa: epidemiology and control strategies [J]. Can J Plant Pathol, 2003, 25(4): 328-338. |
| [15] | Bian XJ, Jing FX, Li G, et al. A microfluidic droplet digital PCR for simultaneous detection of pathogenic Escherichia coli O157 and Listeria monocytogenes [J]. Biosens Bioelectron, 2015, 74: 770-777. |
| [16] | Moukhamedov R. Use of polymerase chain reaction-amplified ribosomal intergenic sequences for the diagnosis of Verticillium tricorpus [J]. Phytopathology, 1994, 84(3): 256. |
| [17] | 朱荷琴, 李志芳, 冯自力, 等. 我国棉花黄萎病研究十年回顾及展望 [J]. 棉花学报, 2017, 29(S1): 37-50. |
| Zhu HQ, Li ZF, Feng ZL, et al. Overview of cotton Verticillium wilt research over the past decade in China and its prospect in future [J]. Cotton Science, 2017, 29(S1): 37-50. | |
| [18] | Gayoso C, de Ilárduya OM, Pomar F, et al. Assessment of real-time PCR as a method for determining the presence of Verticillium dahliae in different Solanaceae cultivars [J]. Eur J Plant Pathol, 2007, 118(3): 199-209. |
| [19] | Bustin SA, Benes V, Garson JA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments [J]. Clin Chem, 2009, 55(4): 611-622. |
| [20] | Mai GQ, Chen JY, Zhang M, et al. Construction of a pathogenic microorganism detection method based on third-generation nanopore sequencing data [J]. BMC Infect Dis, 2025, 25(1): 189. |
| [21] | Huggett JF, Cowen S, Foy CA. Considerations for digital PCR as an accurate molecular diagnostic tool [J]. Clin Chem, 2015, 61(1): 79-88. |
| [22] | Sanders R, Huggett JF, Bushell CA, et al. Evaluation of digital PCR for absolute DNA quantification [J]. Anal Chem, 2011, 83(17): 6474-6484. |
| [23] | Alikian M, Whale AS, Akiki S, et al. RT-qPCR and RT-digital PCR: a comparison of different platforms for the evaluation of residual disease in chronic myeloid leukemia [J]. Clin Chem, 2017, 63(2): 525-531. |
| [24] | Pinheiro LB, Coleman VA, Hindson CM, et al. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification [J]. Anal Chem, 2012, 84(2): 1003-1011. |
| [25] | Hindson CM, Chevillet JR, Briggs HA, et al. Absolute quantification by droplet digital PCR versus analog real-time PCR [J]. Nat Methods, 2013, 10(10): 1003-1005. |
| [26] | Wang D, Liu EL, Liu HY, et al. A droplet digital PCR assay for detection and quantification of Verticillium nonalfalfae and V. albo-atrum [J]. Front Cell Infect Microbiol, 2023, 12: 1110684. |
| [27] | Wang D, Jiao XY, Jia HJ, et al. Detection and quantification of Verticillium dahliae and V. longisporum by droplet digital PCR versus quantitative real-time PCR [J]. Front Cell Infect Microbiol, 2022, 12: 995705. |
| [28] | Moradi A, Almasi MA, Jafary H, et al. A novel and rapid loop-mediated isothermal amplification assay for the specific detection of Verticillium dahliae [J]. J Appl Microbiol, 2014, 116(4): 942-954. |
| [29] | Boogaerts T, Jacobs L, De Roeck N, et al. An alternative approach for bioanalytical assay optimization for wastewater-based epidemiology of SARS-CoV-2 [J]. Sci Total Environ, 2021, 789: 148043. |
| [30] | Ahmad NA, Heng LY, Salam F, et al. A colorimetric pH sensor based on Clitoria sp and Brassica sp for monitoring of food spoilage using chromametry [J]. Sensors, 2019, 19(21): 4813. |
| [31] | Dingle TC, Sedlak RH, Cook L, et al. Tolerance of droplet-digital PCR vs real-time quantitative PCR to inhibitory substances [J]. Clin Chem, 2013, 59(11): 1670-1672. |
| [32] | 黄瑾, 梁涛波, 许恒毅. 数字PCR在生物学检测中应用的研究进展 [J]. 生命科学, 2021, 33(2): 255-264. |
| Huang J, Liang TB, Xu HY. Research progress of application of digital PCR in biological detection [J]. Chinese Bulletin of Life Sciences, 2021, 33(2): 255-264. | |
| [33] | Wang M, Yang JJ, Gai ZT, et al. Comparison between digital PCR and real-time PCR in detection of Salmonella typhimurium in milk [J]. Int J Food Microbiol, 2018, 266: 251-256. |
| [1] | 刘泽洲, 段乃彬, 岳丽昕, 王清华, 姚行浩, 高莉敏, 孔素萍. 大蒜叶片蜡质成分分析及蜡质缺失基因Ggl-1筛选[J]. 生物技术通报, 2025, 41(9): 219-231. |
| [2] | 柴军发, 洪波, 贾彦霞. 转录组和代谢组联合分析三株蜡蚧轮枝菌菌株毒力差异[J]. 生物技术通报, 2025, 41(8): 311-321. |
| [3] | 安苗苗, 蔺祥淏, 梁瑞娜, 赵国柱. 碳源对产甲烷菌群调控及产甲烷能力的影响[J]. 生物技术通报, 2025, 41(6): 355-366. |
| [4] | 周江鸿, 夏菲, 仲丽, 仇兰芬, 李广, 刘倩, 张国锋, 邵金丽, 李娜, 车少臣. 黄栌枯萎病拮抗细菌CCBC3-3-1的全基因组测序及比较基因组分析[J]. 生物技术通报, 2024, 40(7): 235-246. |
| [5] | 余慧, 王静, 梁昕昕, 辛亚平, 周军, 赵会君. 宁夏枸杞铁镉响应基因的筛选及其功能验证[J]. 生物技术通报, 2023, 39(7): 195-205. |
| [6] | 潘国强, 吴思源, 刘璐, 郭惠明, 程红梅, 苏晓峰. 大丽轮枝菌(Verticillim dahliae)突变体库的构建与分析[J]. 生物技术通报, 2023, 39(5): 112-119. |
| [7] | 姚姿婷, 曹雪颖, 肖雪, 李瑞芳, 韦小妹, 邹承武, 朱桂宁. 火龙果溃疡病菌实时荧光定量PCR内参基因的筛选[J]. 生物技术通报, 2023, 39(5): 92-102. |
| [8] | 宋海娜, 吴心桐, 杨鲁豫, 耿喜宁, 张华敏, 宋小龙. 葱鳞葡萄孢菌诱导下韭菜RT-qPCR内参基因的筛选和验证[J]. 生物技术通报, 2023, 39(3): 101-115. |
| [9] | 穆德添, 万凌云, 章瑶, 韦树根, 陆英, 付金娥, 田艺, 潘丽梅, 唐其. 钩藤管家基因筛选及生物碱合成相关基因的表达分析[J]. 生物技术通报, 2023, 39(2): 126-138. |
| [10] | 李会杰, 董莲华, 陈桂芳, 刘思渊, 杨佳怡, 杨靖亚. 食品中椰毒假单胞菌微滴式数字PCR定量检测方法的建立[J]. 生物技术通报, 2023, 39(1): 127-136. |
| [11] | 胡雪莹, 张越, 郭雅杰, 仇天雷, 高敏, 孙兴滨, 王旭明. 不同施肥处理农田土壤中噬菌体与细菌携带抗生素抗性基因的比较[J]. 生物技术通报, 2022, 38(9): 116-126. |
| [12] | 程深伟, 张克强, 梁军锋, 刘福元, 郜兴亮, 杜连柱. 畜禽养殖粪污中典型致病菌的三重微滴式数字PCR定量检测方法的建立[J]. 生物技术通报, 2022, 38(9): 271-280. |
| [13] | 曹英芳, 赵新, 刘双, 李瑞环, 刘娜, 徐石勇, 高芳瑞, 马卉, 兰青阔, 檀建新, 王永. 抗除草剂大豆GE-J12实时荧光定量PCR检测方法的建立[J]. 生物技术通报, 2022, 38(7): 146-152. |
| [14] | 徐圆圆, 赵国春, 郝颖颖, 翁学煌, 陈仲, 贾黎明. 无患子RT-qPCR内参基因的筛选与验证[J]. 生物技术通报, 2022, 38(10): 80-89. |
| [15] | 田李, 李俊娇, 戴小枫, 张丹丹, 陈捷胤. 从功能基因到生物学性状:大丽轮枝菌致病性形成的分子基础[J]. 生物技术通报, 2022, 38(1): 51-69. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||