








生物技术通报 ›› 2023, Vol. 39 ›› Issue (5): 92-102.doi: 10.13560/j.cnki.biotech.bull.1985.2022-1322
姚姿婷1( ), 曹雪颖2, 肖雪2, 李瑞芳1, 韦小妹1, 邹承武2(
), 曹雪颖2, 肖雪2, 李瑞芳1, 韦小妹1, 邹承武2( ), 朱桂宁1(
), 朱桂宁1( )
)
收稿日期:2022-10-26
出版日期:2023-05-26
发布日期:2023-06-08
通讯作者:
邹承武,男,博士,讲师,研究方向:植物病原真菌致病机理;E-mail:zouchengwu@163.com;作者简介:姚姿婷,女,博士,副研究员,研究方向:特色果蔬病害发生与综合防治;E-mail:youziting@163.com
基金资助:
YAO Zi-ting1( ), CAO Xue-ying2, XIAO Xue2, LI Rui-fang1, WEI Xiao-mei1, ZOU Cheng-wu2(
), CAO Xue-ying2, XIAO Xue2, LI Rui-fang1, WEI Xiao-mei1, ZOU Cheng-wu2( ), ZHU Gui-ning1(
), ZHU Gui-ning1( )
)
Received:2022-10-26
Published:2023-05-26
Online:2023-06-08
摘要:
火龙果溃疡病是由火龙果溃疡病菌引起的重要真菌病害,近年来在世界火龙果产区的爆发严重影响了火龙果产业的发展。为研究火龙果溃疡病菌响应逆境胁迫的基因表达和功能,需要筛选火龙果溃疡病菌在各种条件下表达稳定的内参基因。以火龙果溃疡病菌菌株LJ02为研究对象,以在马铃薯葡萄糖液体培养基(PDW)培养的菌丝体为对照组,以在LB培养基培养的菌丝体和分别在添加过氧化氢、吡唑醚菌酯和苯醚甲环唑的PDW培养的菌丝体为处理组。利用实时荧光定量PCR技术以及内参基因稳定性评估软件(geNorm、NormFinder、Bestkeeper和RefFinder),评估18 个候选内参基因(CYT1、SDH2_1、TBCC、SUI1、RPL19、PPH、ATP5B、UBE2_16、RPL13、UBE2_2、PRS17、SUCLA2、ATP5A、TUB1_2、ACT1、EFTU、EF1A和GAPDH)的表达稳定性。结果显示,火龙果溃疡病菌的18个候选基因在上述培养条件下的Ct值介于14.80-24.66之间,表达水平适中;稳定性最高的内参基因是CYT1,其次为SUI1,GAPDH和ACT1的稳定性最差;最少使用2个内参基因组合可以提高定量PCR分析结果的准确性,此时SUCLA2和ATP5A是最稳定的内参基因组合。该结果可为研究火龙果溃疡病菌生物学过程中的基因表达提供理论依据。
姚姿婷, 曹雪颖, 肖雪, 李瑞芳, 韦小妹, 邹承武, 朱桂宁. 火龙果溃疡病菌实时荧光定量PCR内参基因的筛选[J]. 生物技术通报, 2023, 39(5): 92-102.
YAO Zi-ting, CAO Xue-ying, XIAO Xue, LI Rui-fang, WEI Xiao-mei, ZOU Cheng-wu, ZHU Gui-ning. Screening of Reference Genes for RT-qPCR in Neoscytalidium dimidiatum[J]. Biotechnology Bulletin, 2023, 39(5): 92-102.
| 序号 No. | 基因ID Gene ID | 引物 Primer | 引物序列 Primer sequence(5'-3') | 退火温度 Annealing temperature/℃ | 产物长度 PCR product length/bp | 基因名称 Gene name | 基因注释 Gene description |
|---|---|---|---|---|---|---|---|
| Gene1 | NEO_001463 | gene1-qF | GCGGTTTCCAGGTCTATCGTGAAG | 59 | 139 | CYT1 | Cytochrome c1 |
| gene1-qR | GGCTCGGTGTCGTACTCGTTCT | ||||||
| Gene2 | NEO_001659 | gene2-qF | CGTACTGGTGGAACTCGGAGGA | 58.5 | 151 | SDH2_1 | Succinate dehydrogenase subunit B |
| gene2-qR | TTGAGAATGGTGTGGCAACGGTAA | ||||||
| Gene3 | NEO_002342 | gene3-qF | GCTCGCCAATCTCCGTCGTT | 61.1 | 176 | TBCC | Tubulin-specific chaperone c |
| gene3-qR | TCACCACCAGCACGCTATCCT | ||||||
| Gene4 | NEO_002585 | gene4-qF | AGACACAGGCGAGACCAAGCA | 55.8 | 151 | SUI1 | Translation initiation factor SUI1 |
| gene4-qR | GCAGGCAAACTTCTTCTTGATGACC | ||||||
| Gene5 | NEO_003941 | gene5-qF | ATGCCGAGCCAGAAGTCTTTCC | 57.9 | 117 | RPL39 | Ribosomal L39 protein |
| gene5-qR | GGCGTTGTACCTGATGGTGTTG | ||||||
| Gene6 | NEO_004521 | gene6-qF | AAGTCTATGGCTTCTACGACGAGTG | 58.4 | 198 | PPH | Serine threonine-protein phosphatase pp2a catalytic subunit |
| gene6-qR | GCGGCACCTCTTGTATCCTGTC | ||||||
| Gene7 | NEO_004849 | gene7-qF | GCCACCAAGTTCGCTCCTATCC | 58.4 | 147 | ATP5B | ATP synthase F1 beta subunit |
| gene7-qR | GGCACCACCGAAGAGACCAATC | ||||||
| Gene8 | NEO_006464 | gene8-qF | CGGTGTCTTCTTCCTCGCTATCC | 58.3 | 183 | UBE2_16 | Ubiquitin-conjugating enzyme E2-16 kD |
| gene8-qR | GAGCAGATGGAGAGCAGAACCTTC | ||||||
| Gene9 | NEO_008113 | gene9-qF | TCACGAGTTCGGCTGGAAGTACC | 59.7 | 165 | RPL13 | Ribosomal L13 protein |
| gene9-qR | TCGGCAAGCTGAGTCTTGACCTT | ||||||
| Gene10 | NEO_008284 | gene10-qF | TCTGCTCAACGACCCGAACAC | 56.4 | 115 | UBE2_2 | Ubiquitin-conjugating enzyme e2_2 |
| gene10-qR | CCAACTCTTCTCCACCGTCTCC | ||||||
| Gene11 | NEO_008330 | gene11-qF | CTGTCAAGAAGTCCGCCAAGGT | 56.5 | 157 | RPS17 | Ribosomal protein S17e |
| gene11-qR | CGCTTCATCAAGTGAGTGGTGTAG | ||||||
| Gene12 | NEO_008873 | gene12-qF | TGGTGACATTGGCTGCCTGGT | 60.4 | 186 | SUCLA2 | Succinyl-CoA synthetase beta subunit |
| gene12-qR | ATGTTGACGAAGATGGCGGTGAC | ||||||
| Gene13 | NEO_010290 | gene13-qF | TCCTCTCATCTTCGCTGGTGTCA | 56.2 | 110 | ATP5A | ATP synthase F1 alpha subunit |
| gene13-qR | GCTCGGACTCGTTGCTCTTCAG | ||||||
| Gene14 | NEO_010575 | gene14-qF | CAAGGAGGATGCCGCCAACAA | 59 | 145 | TUB1_2 | Tubulin alpha-2 chain |
| gene14-qR | ACCGCCGAAGGAGTGGAAGAT | ||||||
| Gene15 | NEO_011231 | gene15-qF | CCAAGTCCAACCGTGAGAAGATGA | 56.7 | 102 | ACT1 | Actin |
| gene15-qR | CGGAAGCGTACAGCGACAGAA | ||||||
| Gene16 | NEO_011865 | gene16-qF | AGACCGACAACCGCCACTATGC | 60.1 | 184 | EFTU | Translation elongation factor Tu |
| gene16-qR | ACAATCTTCTGGACACCGACCTGAC | ||||||
| Gene17 | NEO_012815 | gene17-qF | CTGGTGAGTTCGAGGCTGGTATCT | 58.2 | 121 | EF1A | Translation elongation factor 1-alpha protein |
| gene17-qR | CACTTGGTGGTGTCCATCTTGTTGA | ||||||
| Gene18 | NEO_013111 | gene18-qF | CCGTACCGCTGCTACCAACATC | 59.8 | 195 | GAPDH | Glyceraldehyde-3-phosphate dehydrogenase |
| gene18-qR | GCCTTGATAGTCGCCTTGATCTCAT |
表1 候选内参基因的引物序列
Table 1 Primer sequences of candidate reference genes
| 序号 No. | 基因ID Gene ID | 引物 Primer | 引物序列 Primer sequence(5'-3') | 退火温度 Annealing temperature/℃ | 产物长度 PCR product length/bp | 基因名称 Gene name | 基因注释 Gene description |
|---|---|---|---|---|---|---|---|
| Gene1 | NEO_001463 | gene1-qF | GCGGTTTCCAGGTCTATCGTGAAG | 59 | 139 | CYT1 | Cytochrome c1 |
| gene1-qR | GGCTCGGTGTCGTACTCGTTCT | ||||||
| Gene2 | NEO_001659 | gene2-qF | CGTACTGGTGGAACTCGGAGGA | 58.5 | 151 | SDH2_1 | Succinate dehydrogenase subunit B |
| gene2-qR | TTGAGAATGGTGTGGCAACGGTAA | ||||||
| Gene3 | NEO_002342 | gene3-qF | GCTCGCCAATCTCCGTCGTT | 61.1 | 176 | TBCC | Tubulin-specific chaperone c |
| gene3-qR | TCACCACCAGCACGCTATCCT | ||||||
| Gene4 | NEO_002585 | gene4-qF | AGACACAGGCGAGACCAAGCA | 55.8 | 151 | SUI1 | Translation initiation factor SUI1 |
| gene4-qR | GCAGGCAAACTTCTTCTTGATGACC | ||||||
| Gene5 | NEO_003941 | gene5-qF | ATGCCGAGCCAGAAGTCTTTCC | 57.9 | 117 | RPL39 | Ribosomal L39 protein |
| gene5-qR | GGCGTTGTACCTGATGGTGTTG | ||||||
| Gene6 | NEO_004521 | gene6-qF | AAGTCTATGGCTTCTACGACGAGTG | 58.4 | 198 | PPH | Serine threonine-protein phosphatase pp2a catalytic subunit |
| gene6-qR | GCGGCACCTCTTGTATCCTGTC | ||||||
| Gene7 | NEO_004849 | gene7-qF | GCCACCAAGTTCGCTCCTATCC | 58.4 | 147 | ATP5B | ATP synthase F1 beta subunit |
| gene7-qR | GGCACCACCGAAGAGACCAATC | ||||||
| Gene8 | NEO_006464 | gene8-qF | CGGTGTCTTCTTCCTCGCTATCC | 58.3 | 183 | UBE2_16 | Ubiquitin-conjugating enzyme E2-16 kD |
| gene8-qR | GAGCAGATGGAGAGCAGAACCTTC | ||||||
| Gene9 | NEO_008113 | gene9-qF | TCACGAGTTCGGCTGGAAGTACC | 59.7 | 165 | RPL13 | Ribosomal L13 protein |
| gene9-qR | TCGGCAAGCTGAGTCTTGACCTT | ||||||
| Gene10 | NEO_008284 | gene10-qF | TCTGCTCAACGACCCGAACAC | 56.4 | 115 | UBE2_2 | Ubiquitin-conjugating enzyme e2_2 |
| gene10-qR | CCAACTCTTCTCCACCGTCTCC | ||||||
| Gene11 | NEO_008330 | gene11-qF | CTGTCAAGAAGTCCGCCAAGGT | 56.5 | 157 | RPS17 | Ribosomal protein S17e |
| gene11-qR | CGCTTCATCAAGTGAGTGGTGTAG | ||||||
| Gene12 | NEO_008873 | gene12-qF | TGGTGACATTGGCTGCCTGGT | 60.4 | 186 | SUCLA2 | Succinyl-CoA synthetase beta subunit |
| gene12-qR | ATGTTGACGAAGATGGCGGTGAC | ||||||
| Gene13 | NEO_010290 | gene13-qF | TCCTCTCATCTTCGCTGGTGTCA | 56.2 | 110 | ATP5A | ATP synthase F1 alpha subunit |
| gene13-qR | GCTCGGACTCGTTGCTCTTCAG | ||||||
| Gene14 | NEO_010575 | gene14-qF | CAAGGAGGATGCCGCCAACAA | 59 | 145 | TUB1_2 | Tubulin alpha-2 chain |
| gene14-qR | ACCGCCGAAGGAGTGGAAGAT | ||||||
| Gene15 | NEO_011231 | gene15-qF | CCAAGTCCAACCGTGAGAAGATGA | 56.7 | 102 | ACT1 | Actin |
| gene15-qR | CGGAAGCGTACAGCGACAGAA | ||||||
| Gene16 | NEO_011865 | gene16-qF | AGACCGACAACCGCCACTATGC | 60.1 | 184 | EFTU | Translation elongation factor Tu |
| gene16-qR | ACAATCTTCTGGACACCGACCTGAC | ||||||
| Gene17 | NEO_012815 | gene17-qF | CTGGTGAGTTCGAGGCTGGTATCT | 58.2 | 121 | EF1A | Translation elongation factor 1-alpha protein |
| gene17-qR | CACTTGGTGGTGTCCATCTTGTTGA | ||||||
| Gene18 | NEO_013111 | gene18-qF | CCGTACCGCTGCTACCAACATC | 59.8 | 195 | GAPDH | Glyceraldehyde-3-phosphate dehydrogenase |
| gene18-qR | GCCTTGATAGTCGCCTTGATCTCAT |
图1 总RNA 凝胶电泳图 M:1 kb plus marker;1-3:对照组3个重复;4-6: A组3个重复;7-9: B组3个重复;10-12: C组3个重复;13-15: D组3个重复
Fig. 1 Electrophoresis analysis of total RNA M: 1 kb plus marker; 1-3: 3 biological repeats of control group; 4-6: 3 biological repeats of A group; 7-9: 3 biological repeats of B group; 10-12: 3 biological repeats of C group; 13-15: 3 biological repeats of D group

图2 18个候选内参基因扩增产物的凝胶电泳分析
Fig. 2 Electrophoretic analysis of amplified products of 18 candidate reference genes M: 1 kb plus marker; nc: blank control; 1: CYT1; 2: SDH2_1; 3: TBCC; 4: SUI1;5: RPL39; 6: PPH; 7: ATP5B; 8: UBE2_16; 9: RPL13; 10: UBE2_2; 11: RPS17; 12: SUCLA2; 13: ATP5A; 14: TUB1_2; 15: ACT1; 16: EFTU; 17: EF1A; 18: GAPDH
| 序号 No. | 内参基因 Reference gene | 扩增效率 RT-qPCR efficiency/% | 相关系数 Correlation coefficient R2 | 序号 No. | 内参基因 Reference gene | 扩增效率 RT-qPCR efficiency/% | 相关系数 Correlation coefficient R2 | |
|---|---|---|---|---|---|---|---|---|
| Gene 1 | CYT1 | 106.4 | 0.998 5 | Gene 10 | UBE2_2 | 96.5 | 0.999 9 | |
| Gene 2 | SDH2_1 | 106.2 | 0.986 5 | Gene 11 | RPS17 | 96.7 | 0.996 6 | |
| Gene 3 | TBCC | 104.3 | 0.989 4 | Gene 12 | SUCLA2 | 103.6 | 0.996 6 | |
| Gene 4 | SUI1 | 100.1 | 0.999 5 | Gene 13 | ATP5A | 101.9 | 0.996 9 | |
| Gene 5 | RPL39 | 108.3 | 0.998 8 | Gene 14 | TUB1_2 | 93.6 | 0.999 6 | |
| Gene 6 | PPH | 108.9 | 0.999 8 | Gene 15 | ACT1 | 91.6 | 0.999 9 | |
| Gene 7 | ATP5B | 104.7 | 0.997 4 | Gene 16 | EFTU | 102.4 | 0.994 1 | |
| Gene 8 | UBE2_16 | 104.2 | 0.999 9 | Gene 17 | EF1A | 94.8 | 0.996 8 | |
| Gene 9 | RPL13 | 100.9 | 0.999 9 | Gene 18 | GAPDH | 90.9 | 0.995 0 |
表2 候选内参基因的扩增效率
Table 2 Amplification efficiencies of candidate reference genes
| 序号 No. | 内参基因 Reference gene | 扩增效率 RT-qPCR efficiency/% | 相关系数 Correlation coefficient R2 | 序号 No. | 内参基因 Reference gene | 扩增效率 RT-qPCR efficiency/% | 相关系数 Correlation coefficient R2 | |
|---|---|---|---|---|---|---|---|---|
| Gene 1 | CYT1 | 106.4 | 0.998 5 | Gene 10 | UBE2_2 | 96.5 | 0.999 9 | |
| Gene 2 | SDH2_1 | 106.2 | 0.986 5 | Gene 11 | RPS17 | 96.7 | 0.996 6 | |
| Gene 3 | TBCC | 104.3 | 0.989 4 | Gene 12 | SUCLA2 | 103.6 | 0.996 6 | |
| Gene 4 | SUI1 | 100.1 | 0.999 5 | Gene 13 | ATP5A | 101.9 | 0.996 9 | |
| Gene 5 | RPL39 | 108.3 | 0.998 8 | Gene 14 | TUB1_2 | 93.6 | 0.999 6 | |
| Gene 6 | PPH | 108.9 | 0.999 8 | Gene 15 | ACT1 | 91.6 | 0.999 9 | |
| Gene 7 | ATP5B | 104.7 | 0.997 4 | Gene 16 | EFTU | 102.4 | 0.994 1 | |
| Gene 8 | UBE2_16 | 104.2 | 0.999 9 | Gene 17 | EF1A | 94.8 | 0.996 8 | |
| Gene 9 | RPL13 | 100.9 | 0.999 9 | Gene 18 | GAPDH | 90.9 | 0.995 0 |

图3 候选内参基因的熔解曲线分析
Fig. 3 Melting curve analysis of candidate reference genes Gene 1: CYT1; Gene 2: SDH2_1; Gene 3: TBCC; Gene 4: SUI1; Gene 5: RPL39; Gene 6: PPH; Gene 7: ATP5B; Gene 8: UBE2_16; Gene 9: RPL13; Gene 10: UBE2_2; Gene 11: RPS17; Gene 12: SUCLA2; Gene 13: ATP5A; Gene 14: TUB1_2; Gene 15: ACT1; Gene 16: EFTU; Gene 17: EF1A; Gene 18: GAPDH. The same below
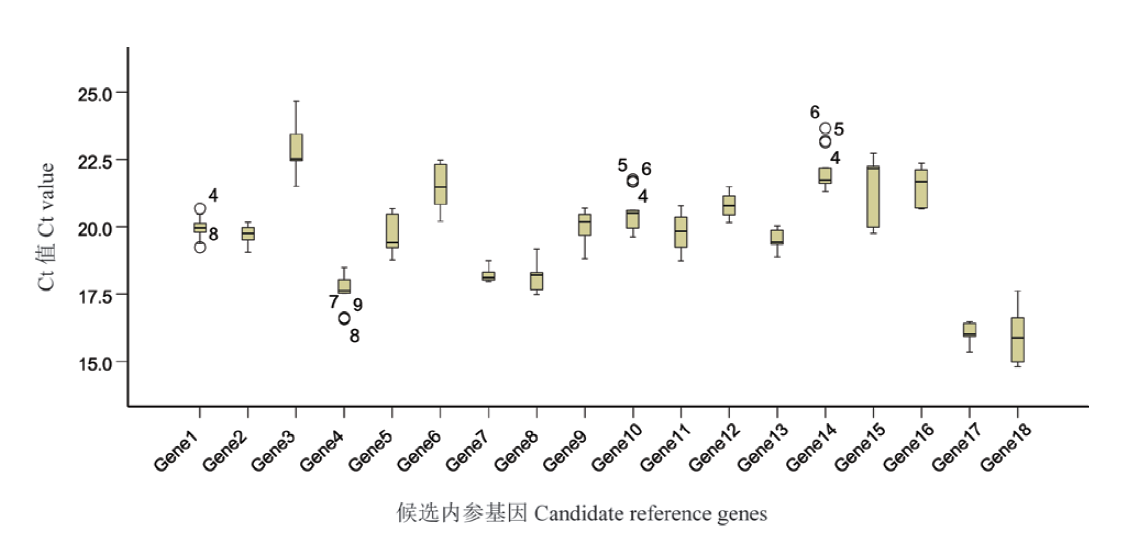
图4 不同培养条件下各个候选参考基因的表达水平 箱型图表示Ct值范围的上下四分位数及中位数,以及上下边缘代表最大值和最小值
Fig. 4 Expression levels of candidate reference genes in different culture conditions Boxes represent lower and upper quartiles of cycle thresholds range with medians indicated, and whisker caps represent maximum and minimum values
| 基因序号 No. | 基因名称 Gene name | 几何平均值 Geo mean | 算术平均值 AR mean | 最小值 Min | 最大值 Max | 相关系数 Correlation coefficient(r) | 标准偏差 Std dev | 变异系数 CV |
|---|---|---|---|---|---|---|---|---|
| Gene1 | CYT1 | 19.94 | 19.94 | 19.24 | 20.68 | 0.960 | 0.31 | 1.53 |
| Gene2 | SDH2_1 | 19.69 | 19.69 | 19.06 | 20.16 | 0.754 | 0.30 | 1.54 |
| Gene3 | TBCC | 22.90 | 22.92 | 21.50 | 24.66 | 0.915 | 0.83 | 3.63 |
| Gene4 | SUI1 | 17.65 | 17.66 | 16.58 | 18.49 | 0.952 | 0.47 | 2.66 |
| Gene5 | RPL39 | 19.71 | 19.72 | 18.77 | 20.67 | 0.851 | 0.63 | 3.21 |
| Gene6 | PPH | 21.46 | 21.47 | 20.20 | 22.48 | 0.936 | 0.72 | 3.38 |
| Gene7 | ATP5B | 18.21 | 18.22 | 17.97 | 18.74 | 0.521 | 0.20 | 1.11 |
| Gene8 | UBE2_16 | 18.16 | 18.17 | 17.48 | 19.18 | 0.912 | 0.44 | 2.40 |
| Gene9 | RPL13 | 19.97 | 19.98 | 18.81 | 20.69 | 0.761 | 0.56 | 2.79 |
| Gene10 | UBE2_2 | 20.47 | 20.49 | 19.63 | 21.76 | 0.963 | 0.53 | 2.6 |
| Gene11 | RPS17 | 19.82 | 19.83 | 18.73 | 20.78 | 0.803 | 0.61 | 3.09 |
| Gene12 | SUCLA2 | 20.78 | 20.79 | 20.15 | 21.48 | 0.825 | 0.38 | 1.81 |
| Gene13 | ATP5A | 19.49 | 19.50 | 18.88 | 20.03 | 0.87 | 0.33 | 1.67 |
| Gene14 | TUB1_2 | 22.06 | 22.07 | 21.31 | 23.66 | 0.892 | 0.55 | 2.50 |
| Gene15 | ACT1 | 21.30 | 21.33 | 19.76 | 22.72 | 0.835 | 1.13 | 5.29 |
| Gene16 | EFTU | 21.48 | 21.49 | 20.66 | 22.36 | 0.577 | 0.65 | 3.00 |
| Gene17 | EF1A | 16.04 | 16.05 | 15.35 | 16.48 | 0.893 | 0.31 | 1.96 |
| Gene18 | GAPDH | 15.97 | 16.00 | 14.80 | 17.62 | 0.929 | 0.89 | 5.56 |
表3 BestKeeper 方法评价候选内参基因稳定性
Table 3 Evaluating the expression stabilities of candidate reference genes by BestKeeper
| 基因序号 No. | 基因名称 Gene name | 几何平均值 Geo mean | 算术平均值 AR mean | 最小值 Min | 最大值 Max | 相关系数 Correlation coefficient(r) | 标准偏差 Std dev | 变异系数 CV |
|---|---|---|---|---|---|---|---|---|
| Gene1 | CYT1 | 19.94 | 19.94 | 19.24 | 20.68 | 0.960 | 0.31 | 1.53 |
| Gene2 | SDH2_1 | 19.69 | 19.69 | 19.06 | 20.16 | 0.754 | 0.30 | 1.54 |
| Gene3 | TBCC | 22.90 | 22.92 | 21.50 | 24.66 | 0.915 | 0.83 | 3.63 |
| Gene4 | SUI1 | 17.65 | 17.66 | 16.58 | 18.49 | 0.952 | 0.47 | 2.66 |
| Gene5 | RPL39 | 19.71 | 19.72 | 18.77 | 20.67 | 0.851 | 0.63 | 3.21 |
| Gene6 | PPH | 21.46 | 21.47 | 20.20 | 22.48 | 0.936 | 0.72 | 3.38 |
| Gene7 | ATP5B | 18.21 | 18.22 | 17.97 | 18.74 | 0.521 | 0.20 | 1.11 |
| Gene8 | UBE2_16 | 18.16 | 18.17 | 17.48 | 19.18 | 0.912 | 0.44 | 2.40 |
| Gene9 | RPL13 | 19.97 | 19.98 | 18.81 | 20.69 | 0.761 | 0.56 | 2.79 |
| Gene10 | UBE2_2 | 20.47 | 20.49 | 19.63 | 21.76 | 0.963 | 0.53 | 2.6 |
| Gene11 | RPS17 | 19.82 | 19.83 | 18.73 | 20.78 | 0.803 | 0.61 | 3.09 |
| Gene12 | SUCLA2 | 20.78 | 20.79 | 20.15 | 21.48 | 0.825 | 0.38 | 1.81 |
| Gene13 | ATP5A | 19.49 | 19.50 | 18.88 | 20.03 | 0.87 | 0.33 | 1.67 |
| Gene14 | TUB1_2 | 22.06 | 22.07 | 21.31 | 23.66 | 0.892 | 0.55 | 2.50 |
| Gene15 | ACT1 | 21.30 | 21.33 | 19.76 | 22.72 | 0.835 | 1.13 | 5.29 |
| Gene16 | EFTU | 21.48 | 21.49 | 20.66 | 22.36 | 0.577 | 0.65 | 3.00 |
| Gene17 | EF1A | 16.04 | 16.05 | 15.35 | 16.48 | 0.893 | 0.31 | 1.96 |
| Gene18 | GAPDH | 15.97 | 16.00 | 14.80 | 17.62 | 0.929 | 0.89 | 5.56 |
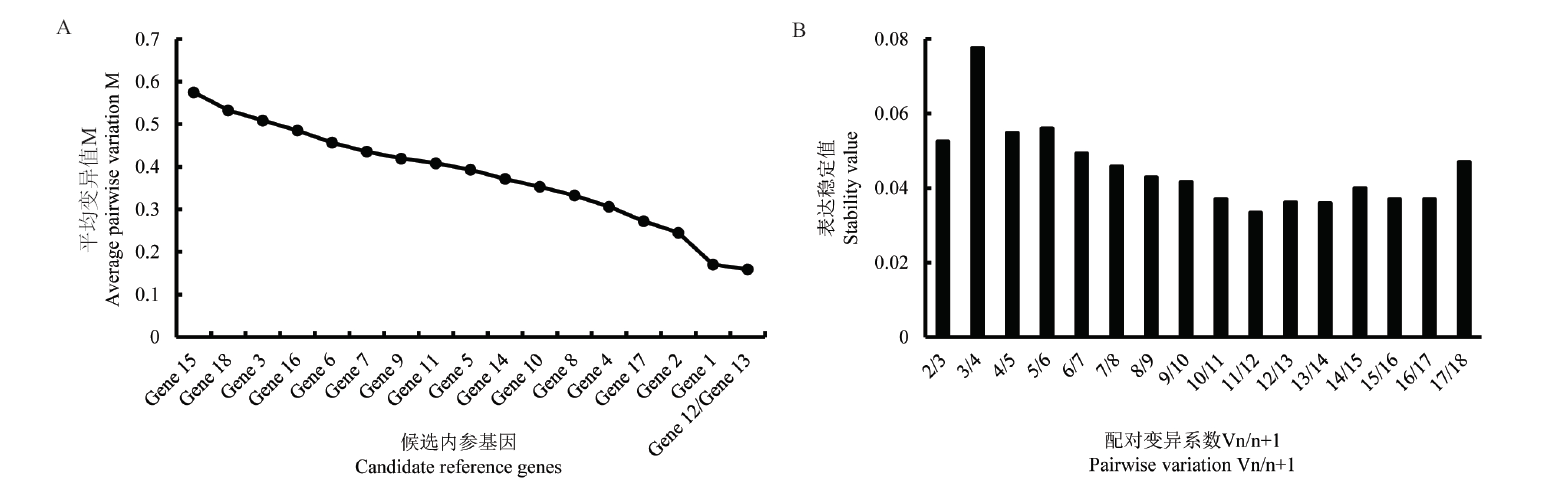
图5 geNorm分析18个候选参考基因的表达稳定性(A)和内参基因最适数目(B)
Fig. 5 Analyzing the expression stabilities(A)and optimal numbers(B)of 18 candidate reference genes by geNorm
| 序号 No. | 基因 Genes | geNorm | NormFinder | BestKeeper | RefFinder | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M值 M value | 排序 Rank | 稳定值 Stability value | 排序 Rank | 变异度 CV/% | 标准差 SD | 相关系数 Correlation coefficientr | 排序 Rank | 稳定性 Stability value | 综合排序 Comprehensive rank | ||||||
| Gene1 | CYT1 | 0.171 | 2 | 0.186 | 2 | 1.53 | 0.31 | 0.960 | 1 | 2.06 | 1 | ||||
| Gene2 | SDH2_1 | 0.245 | 3 | 0.383 | 9 | 1.54 | 0.30 | 0.754 | 13 | 5.05 | 7 | ||||
| Gene3 | TBCC | 0.509 | 15 | 0.563 | 15 | 3.63 | 0.83 | 0.915 | 7 | 15.49 | 15 | ||||
| Gene4 | SUI1 | 0.306 | 5 | 0.185 | 1 | 2.66 | 0.47 | 0.952 | 3 | 3.13 | 2 | ||||
| Gene5 | RPL39 | 0.393 | 9 | 0.391 | 10 | 3.21 | 0.63 | 0.851 | 10 | 10.68 | 11 | ||||
| Gene6 | PPH | 0.457 | 13 | 0.392 | 11 | 3.38 | 0.72 | 0.936 | 5 | 12.63 | 14 | ||||
| Gene7 | ATP5B | 0.436 | 12 | 0.499 | 14 | 1.11 | 0.2 | 0.521 | 16 | 7.10 | 9 | ||||
| Gene8 | UBE2_16 | 0.333 | 6 | 0.232 | 4 | 2.40 | 0.44 | 0.912 | 4 | 5.29 | 8 | ||||
| Gene9 | RPL13 | 0.420 | 11 | 0.462 | 13 | 2.79 | 0.56 | 0.761 | 14 | 12.22 | 13 | ||||
| Gene10 | UBE2_2 | 0.353 | 7 | 0.225 | 3 | 2.60 | 0.53 | 0.963 | 2 | 5.05 | 6 | ||||
| Gene11 | RPS17 | 0.408 | 10 | 0.449 | 12 | 3.09 | 0.61 | 0.803 | 15 | 11.74 | 12 | ||||
| Gene12 | SUCLA2 | 0.159 | 1 | 0.312 | 7 | 1.81 | 0.38 | 0.825 | 12 | 4.14 | 4 | ||||
| Gene13 | ATP5A | 0.159 | 1 | 0.286 | 6 | 1.67 | 0.33 | 0.870 | 8 | 3.50 | 3 | ||||
| Gene14 | TUB1_2 | 0.372 | 8 | 0.341 | 8 | 2.50 | 0.55 | 0.892 | 9 | 8.71 | 10 | ||||
| Gene15 | ACT1 | 0.576 | 17 | 0.839 | 18 | 5.29 | 1.13 | 0.835 | 18 | 18.00 | 18 | ||||
| Gene16 | EFTU | 0.485 | 14 | 0.622 | 17 | 3.00 | 0.65 | 0.577 | 17 | 15.70 | 16 | ||||
| Gene17 | EF1A | 0.272 | 4 | 0.276 | 5 | 1.96 | 0.31 | 0.893 | 6 | 4.95 | 5 | ||||
| Gene18 | GAPDH | 0.533 | 16 | 0.588 | 16 | 5.56 | 0.89 | 0.929 | 11 | 16.49 | 17 | ||||
表4 GeNorm、NormFinder、BestKeeper和RefFinder分析基因表达稳定性结果
Table 4 Analyzing the expression stabilities of genes by GeNorm, NormFinder, BestKeeper and RefFinder
| 序号 No. | 基因 Genes | geNorm | NormFinder | BestKeeper | RefFinder | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M值 M value | 排序 Rank | 稳定值 Stability value | 排序 Rank | 变异度 CV/% | 标准差 SD | 相关系数 Correlation coefficientr | 排序 Rank | 稳定性 Stability value | 综合排序 Comprehensive rank | ||||||
| Gene1 | CYT1 | 0.171 | 2 | 0.186 | 2 | 1.53 | 0.31 | 0.960 | 1 | 2.06 | 1 | ||||
| Gene2 | SDH2_1 | 0.245 | 3 | 0.383 | 9 | 1.54 | 0.30 | 0.754 | 13 | 5.05 | 7 | ||||
| Gene3 | TBCC | 0.509 | 15 | 0.563 | 15 | 3.63 | 0.83 | 0.915 | 7 | 15.49 | 15 | ||||
| Gene4 | SUI1 | 0.306 | 5 | 0.185 | 1 | 2.66 | 0.47 | 0.952 | 3 | 3.13 | 2 | ||||
| Gene5 | RPL39 | 0.393 | 9 | 0.391 | 10 | 3.21 | 0.63 | 0.851 | 10 | 10.68 | 11 | ||||
| Gene6 | PPH | 0.457 | 13 | 0.392 | 11 | 3.38 | 0.72 | 0.936 | 5 | 12.63 | 14 | ||||
| Gene7 | ATP5B | 0.436 | 12 | 0.499 | 14 | 1.11 | 0.2 | 0.521 | 16 | 7.10 | 9 | ||||
| Gene8 | UBE2_16 | 0.333 | 6 | 0.232 | 4 | 2.40 | 0.44 | 0.912 | 4 | 5.29 | 8 | ||||
| Gene9 | RPL13 | 0.420 | 11 | 0.462 | 13 | 2.79 | 0.56 | 0.761 | 14 | 12.22 | 13 | ||||
| Gene10 | UBE2_2 | 0.353 | 7 | 0.225 | 3 | 2.60 | 0.53 | 0.963 | 2 | 5.05 | 6 | ||||
| Gene11 | RPS17 | 0.408 | 10 | 0.449 | 12 | 3.09 | 0.61 | 0.803 | 15 | 11.74 | 12 | ||||
| Gene12 | SUCLA2 | 0.159 | 1 | 0.312 | 7 | 1.81 | 0.38 | 0.825 | 12 | 4.14 | 4 | ||||
| Gene13 | ATP5A | 0.159 | 1 | 0.286 | 6 | 1.67 | 0.33 | 0.870 | 8 | 3.50 | 3 | ||||
| Gene14 | TUB1_2 | 0.372 | 8 | 0.341 | 8 | 2.50 | 0.55 | 0.892 | 9 | 8.71 | 10 | ||||
| Gene15 | ACT1 | 0.576 | 17 | 0.839 | 18 | 5.29 | 1.13 | 0.835 | 18 | 18.00 | 18 | ||||
| Gene16 | EFTU | 0.485 | 14 | 0.622 | 17 | 3.00 | 0.65 | 0.577 | 17 | 15.70 | 16 | ||||
| Gene17 | EF1A | 0.272 | 4 | 0.276 | 5 | 1.96 | 0.31 | 0.893 | 6 | 4.95 | 5 | ||||
| Gene18 | GAPDH | 0.533 | 16 | 0.588 | 16 | 5.56 | 0.89 | 0.929 | 11 | 16.49 | 17 | ||||
| [1] | 吕梦娇, 战春君, 白仲虎, 等. 巴斯德毕赤酵母不同生长阶段内参基因的筛选与验证[J]. 微生物学通报, 2022, 49(11): 4586-4597. |
| Lü MJ, Zhan CJ, Bai ZH, et al. Identification and validation of reference genes for RT-qPCR normalization in Komagataella phaffii at different growth stages[J]. Microbiol China, 2022, 49(11): 4586-4597. | |
| [2] |
Borkowska M, Białas W, Celińska E. A new set of reference genes for comparative gene expression analyses in Yarrowia lipolytica[J]. FEMS Yeast Res, 2020, 20(7): foaa059.
doi: 10.1093/femsyr/foaa059 URL |
| [3] |
Xu ZC, Xu J, Ji AJ, et al. Genome-wide selection of superior reference genes for expression studies in Ganoderma lucidum[J]. Gene, 2015, 574(2): 352-358.
doi: 10.1016/j.gene.2015.08.025 URL |
| [4] |
Ratanaprom S, Nakkanong K, Nualsri C, et al. Overcoming encouragement of dragon fruit plant(Hylocereus undatus)against stem brown spot disease caused by Neoscytalidium dimidiatum using Bacillus subtilis combined with sodium bicarbonate[J]. Plant Pathol J, 2021, 37(3): 205-214.
doi: 10.5423/PPJ.OA.01.2021.0007 pmid: 34111911 |
| [5] | Dy KS, Wonglom P, Pornsuriya C, et al. Morphological, molecular identification and pathogenicity of Neoscytalidium dimidiatum causing stem canker of Hylocereus polyrhizus in southern Thailand[J]. Plants(Basel), 2022, 11(4): 504. |
| [6] | 田婷婷. 马铃薯晚疫病病原菌对H2O2胁迫响应的机制研究[D]. 重庆: 重庆大学, 2020. |
| Tian TT. The response mechanism of potato late blight pathogen to H2O2 stress[D]. Chongqing: Chongqing University, 2020. | |
| [7] | 齐兴柱, 刘磊, 汪军. RNA-Seq揭示Foc4在外源氧化胁迫(H2O2)下的基因表达及细胞代谢变化[J]. 微生物学报, 2019, 59(5): 891-906. |
| Qi XZ, Liu L, Wang J. RNA-Seq reveals changes of gene expression and cellular metabolism caused by exogenous oxidative stress(H2O2)in Foc4[J]. Acta Microbiol Sin, 2019, 59(5): 891-906. | |
| [8] |
Camejo D, Guzmán-Cedeño Á, Moreno A. Reactive oxygen species, essential molecules, during plant-pathogen interactions[J]. Plant Physiol Biochem, 2016, 103: 10-23.
doi: 10.1016/j.plaphy.2016.02.035 URL |
| [9] | 贤小勇, 林珊宇, 朱桂宁, 等. 杀菌剂对火龙果溃疡病的室内毒力和田间防效[J]. 南方农业学报, 2018, 49(7): 1338-1345. |
| Xian XY, Lin SY, Zhu GN, et al. Indoor virulence and field effects of fungicides on pitaya canker[J]. J South Agric, 2018, 49(7): 1338-1345. | |
| [10] | 蓝国兵, 何自福, 于琳, 等. 16种杀菌剂对火龙果褐腐病菌抑菌持效期及田间防效试验[J]. 广东农业科学, 2019, 46(12): 95-101. |
| Lan GB, He ZF, Yu L, et al. Effective inhibition duration and field control effects of 16 fungicides against Neoscytalidium dimidiatum[J]. Guangdong Agric Sci, 2019, 46(12): 95-101. | |
| [11] |
Wan HJ, Zhao ZG, Qian CT, et al. Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber[J]. Anal Biochem, 2010, 399(2): 257-261.
doi: 10.1016/j.ab.2009.12.008 pmid: 20005862 |
| [12] |
Lyu XL, Shen CC, Fu YP, et al. A small secreted virulence-related protein is essential for the necrotrophic interactions of Sclerotinia sclerotiorum with its host plants[J]. PLoS Pathog, 2016, 12(2): e1005435.
doi: 10.1371/journal.ppat.1005435 URL |
| [13] |
Grube S, Göttig T, Freitag D, et al. Selection of suitable reference genes for expression analysis in human glioma using RT-qPCR[J]. J Neurooncol, 2015, 123(1): 35-42.
doi: 10.1007/s11060-015-1772-7 URL |
| [14] |
He YJ, Kim SB, Balint-Kurti P. A maize cytochrome b-c1 complex subunit protein ZmQCR7 controls variation in the hypersensitive response[J]. Planta, 2019, 249(5): 1477-1485.
doi: 10.1007/s00425-019-03092-8 pmid: 30694389 |
| [15] |
Vieira A, Talhinhas P, Loureiro A, et al. Validation of RT-qPCR reference genes for in planta expression studies in Hemileia vastatrix, the causal agent of coffee leaf rust[J]. Fungal Biol, 2011, 115(9): 891-901.
doi: 10.1016/j.funbio.2011.07.002 URL |
| [16] |
Phule AS, Barbadikar KM, Madhav MS, et al. Genes encoding membrane proteins showed stable expression in rice under aerobic condition: novel set of reference genes for expression studies[J]. 3 Biotech, 2018, 8(9): 383.
doi: 10.1007/s13205-018-1406-9 pmid: 30148033 |
| [17] | 周琳琳, 赵玉, 李夏雨, 等. 蚁巢伞属真菌Termitomyces clypeatus实时荧光定量PCR内参基因的筛选[J]. 菌物学报, 2022, 41(10): 1597-1606. |
| Zhou LL, Zhao Y, Li XY, et al. Screening of the reference genes for RT-qPCR analysis of gene expressions in Termitomyces clypeatus[J]. Mycosystema, 2022, 41(10): 1597-1606. | |
| [18] | 盘林秀, 王娜, 王爱军, 等. 稻粒黑粉病菌实时荧光定量PCR内参基因筛选[J]. 植物病理学报, 2018, 48(5): 640-647. |
| Pan LX, Wang N, Wang AJ, et al. Selection of reference genes for quantitative real-time PCR in Tilletia horrida[J]. Acta Phytopathol Sin, 2018, 48(5): 640-647. | |
| [19] | 李兵, 刘柳, 单婷婷, 等. 蜜环菌(Armillaria mellea)内参基因的筛选[J]. 微生物学通报, 2022, 49(2): 473-482. |
| Li B, Liu L, Shan TT, et al. Selection of reference genes for real-time quantitative PCR of Armillaria mellea[J]. Microbiol China, 2022, 49(2): 473-482. | |
| [20] |
Chen XZ, Chen XG, Tan Q, et al. Selection of potential reference genes for RT-qPCR in the plant pathogenic fungus Colletotrichum fructicola[J]. Front Microbiol, 2022, 13: 982748.
doi: 10.3389/fmicb.2022.982748 URL |
| [21] | 苏强军, 夏樱霞, 谢放, 等. 冬虫夏草菌实时荧光定量PCR内参基因的筛选[J]. 菌物学报, 2021, 40(7): 1712-1722. |
| Su QJ, Xia YX, Xie F, et al. Screening of the reference genes for qRT-PCR analysis of gene expression in Ophiocordyceps sinensis[J]. Mycosystema, 2021, 40(7): 1712-1722. | |
| [22] |
Glare EM, Divjak M, Bailey MJ, et al. Beta-Actin and GAPDH housekeeping gene expression in asthmatic airways is variable and not suitable for normalising mRNA levels[J]. Thorax, 2002, 57(9): 765-770.
doi: 10.1136/thorax.57.9.765 pmid: 12200519 |
| [23] |
Tao YX, van Peer AF, Huang QH, et al. Identification of novel and robust internal control genes from Volvariella volvacea that are suitable for RT-qPCR in filamentous fungi[J]. Sci Rep, 2016, 6: 29236.
doi: 10.1038/srep29236 |
| [24] |
Lin JT, Redies C. Histological evidence: housekeeping genes beta-actin and GAPDH are of limited value for normalization of gene expression[J]. Dev Genes Evol, 2012, 222(6): 369-376.
doi: 10.1007/s00427-012-0420-x URL |
| [25] | Yang YY, Xu XY, Jing ZH, et al. Genome-wide screening and stability verification of the robust internal control genes for RT-qPCR in filamentous fungi[J]. J Fungi(Basel), 2022, 8(9): 952. |
| [1] | 王佳蕊, 孙培媛, 柯瑾, 冉彬, 李洪有. 苦荞糖基转移酶基因FtUGT143的克隆及表达分析[J]. 生物技术通报, 2023, 39(8): 204-212. |
| [2] | 孙明慧, 吴琼, 刘丹丹, 焦小雨, 王文杰. 茶树CsTMFs的克隆与表达分析[J]. 生物技术通报, 2023, 39(7): 151-159. |
| [3] | 余慧, 王静, 梁昕昕, 辛亚平, 周军, 赵会君. 宁夏枸杞铁镉响应基因的筛选及其功能验证[J]. 生物技术通报, 2023, 39(7): 195-205. |
| [4] | 赵雪婷, 高利燕, 王俊刚, 沈庆庆, 张树珍, 李富生. 甘蔗AP2/ERF转录因子基因ShERF3的克隆、表达及其编码蛋白的定位[J]. 生物技术通报, 2023, 39(6): 208-216. |
| [5] | 李苑虹, 郭昱昊, 曹燕, 祝振洲, 王飞飞. 外源植物激素调控微藻生长及目标产物积累研究进展[J]. 生物技术通报, 2023, 39(6): 61-72. |
| [6] | 冯珊珊, 王璐, 周益, 王幼平, 方玉洁. WOX家族基因调控植物生长发育和非生物胁迫响应的研究进展[J]. 生物技术通报, 2023, 39(5): 1-13. |
| [7] | 姜晴春, 杜洁, 王嘉诚, 余知和, 王允, 柳忠玉. 虎杖转录因子PcMYB2的表达特性和功能分析[J]. 生物技术通报, 2023, 39(5): 217-223. |
| [8] | 翟莹, 李铭杨, 张军, 赵旭, 于海伟, 李珊珊, 赵艳, 张梅娟, 孙天国. 异源表达大豆转录因子GmNF-YA19提高转基因烟草抗旱性[J]. 生物技术通报, 2023, 39(5): 224-232. |
| [9] | 王艺清, 王涛, 韦朝领, 戴浩民, 曹士先, 孙威江, 曾雯. 茶树SMAS基因家族的鉴定及互作分析[J]. 生物技术通报, 2023, 39(4): 246-258. |
| [10] | 宋海娜, 吴心桐, 杨鲁豫, 耿喜宁, 张华敏, 宋小龙. 葱鳞葡萄孢菌诱导下韭菜RT-qPCR内参基因的筛选和验证[J]. 生物技术通报, 2023, 39(3): 101-115. |
| [11] | 杨春洪, 董璐, 陈林, 宋丽. 大豆VAS1基因家族的鉴定及参与侧根发育的研究[J]. 生物技术通报, 2023, 39(3): 133-142. |
| [12] | 刘思佳, 王浩楠, 付宇辰, 闫文欣, 胡增辉, 冷平生. ‘西伯利亚’百合LiCMK基因克隆及功能分析[J]. 生物技术通报, 2023, 39(3): 196-205. |
| [13] | 王涛, 漆思雨, 韦朝领, 王艺清, 戴浩民, 周喆, 曹士先, 曾雯, 孙威江. CsPPR和CsCPN60-like在茶树白化叶片中的表达分析及互作蛋白验证[J]. 生物技术通报, 2023, 39(3): 218-231. |
| [14] | 庞强强, 孙晓东, 周曼, 蔡兴来, 张文, 王亚强. 菜心BrHsfA3基因克隆及其对高温胁迫的响应[J]. 生物技术通报, 2023, 39(2): 107-115. |
| [15] | 穆德添, 万凌云, 章瑶, 韦树根, 陆英, 付金娥, 田艺, 潘丽梅, 唐其. 钩藤管家基因筛选及生物碱合成相关基因的表达分析[J]. 生物技术通报, 2023, 39(2): 126-138. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||