








生物技术通报 ›› 2022, Vol. 38 ›› Issue (10): 80-89.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1616
徐圆圆1,2,3( ), 赵国春1,2,3, 郝颖颖1,2,3, 翁学煌4, 陈仲1,2,3,5(
), 赵国春1,2,3, 郝颖颖1,2,3, 翁学煌4, 陈仲1,2,3,5( ), 贾黎明1,2,3(
), 贾黎明1,2,3( )
)
收稿日期:2021-12-30
出版日期:2022-10-26
发布日期:2022-11-11
作者简介:徐圆圆,女,博士研究生,研究方向:用材林与能源林培育理论与技术;E-mail:基金资助:
XU Yuan-yuan1,2,3( ), ZHAO Guo-chun1,2,3, HAO Ying-ying1,2,3, WENG Xue-huang4, CHEN Zhong1,2,3,5(
), ZHAO Guo-chun1,2,3, HAO Ying-ying1,2,3, WENG Xue-huang4, CHEN Zhong1,2,3,5( ), JIA Li-ming1,2,3(
), JIA Li-ming1,2,3( )
)
Received:2021-12-30
Published:2022-10-26
Online:2022-11-11
摘要:
无患子根、茎、叶、花和果皮均含有生物活性物质三萜皂苷,为了解三萜皂苷生物合成途径中相关基因的表达水平,需要筛选稳定表达的内参基因。以无患子根、茎、叶、芽、雄花、雌花和不同发育时期的果皮为材料,根据无患子转录组数据,选择Sm18S,SmACT,SmTBCC,SmEF-1α,SmRPL1,SmRPS26,SmUBC12,SmUBP等8个基因作为候选内参基因。通过实时荧光定量PCR(RT-qPCR)检测这些候选内参基因的表达量,并利用geNorm、NormFider和BestKeeper三个软件及RefFinder在线分析工具评价候选内参基因的稳定性。结果表明,8个候选内参基因的表达量在所有样本间的变化幅度存在差异;geNorm、NormFinder和BestKeeper 3个软件筛选出的最佳内参基因略有不同;综合分析结果表明SmACT、SmRPL1和SmUBP表达较为稳定,SmEF-1α稳定性最差;以SmACT、SmACT+SmRPL1组合和SmACT+SmRPL1+SmUBP组合为内参对三萜皂苷生物合成途径中的8个候选基因进行校准所得的表达量基本上保持一致,且相对表达量结果与转录组数据基本保持一致,表明SmACT、SmACT+SmRPL1组合和SmACT+SmRPL1+SmUBP组合可作为无患子三萜皂苷生物合成途径相关基因表达研究的内参基因,同时也可以为无患子及近缘植物的其他生物学过程中的基因表达研究提供参考。
徐圆圆, 赵国春, 郝颖颖, 翁学煌, 陈仲, 贾黎明. 无患子RT-qPCR内参基因的筛选与验证[J]. 生物技术通报, 2022, 38(10): 80-89.
XU Yuan-yuan, ZHAO Guo-chun, HAO Ying-ying, WENG Xue-huang, CHEN Zhong, JIA Li-ming. Reference Genes Selection and Validation for RT-qPCR in Sapindus mukorossi[J]. Biotechnology Bulletin, 2022, 38(10): 80-89.

图1 无患子采样材料 R:根;ST:茎;L:叶;B:芽;MF:雄花;FF:雌花;S1:花后15 d的果皮;S2:花后45 d的果皮;S3:花后75 d的果皮;S4:花后90 d的果皮;S5:花后105 d的果皮;S6:花后120 d的果皮;S7:花后135 d的果皮;S8:花后150 d的果皮
Fig. 1 Sample materials of S. mukorossi R:root;ST:stem;L:leaf;B:bud;MF:male flower;FF:female flower;S1:pericarp at 15 d after flower pollination;S2:pericarp at 45 d after flower pollination;S3:pericarp at 75 d after flower pollination;S4:pericarp at 90 d after flower pollination;S5:pericarp at 105 d after flower pollination;S6:pericarp at 120 d after flower pollination;S7:pericarp at 135 d after flower pollination;S8:pericarp at 150 d after flower pollination
| 内参基因 Reference genes | 基因ID Gene ID | 产物长度 Length of product /bp | 正向引物序列 Forward primer sequences(5'- 3') | 反向引物序列 Reverse primer sequences(5'- 3') |
|---|---|---|---|---|
| Sm18S | Samuk09G0136600 | 221 | AGATTGAAGGCGTTCTTTGGAT | TCAGTGTTTATGGACGGTGGAC |
| SmACT | Samuk13G0061200 | 129 | AGAAAGTTGGCCTCGCTGAA | CAGGAACCAGACCACCTGTC |
| SmEF-1α | Samuk14G0055700 | 101 | TCCAAGGCCAGGTACGATGA | AAACCAGAGATGGGGACGAAG |
| SmRPL1 | Samuk07G0011500 | 206 | CTGACCACCCCACCACTCTC | TCCTCCTCATCTGCCACCTC |
| SmRPS26 | Samuk08G0043900 | 230 | GCGAAGAAATGGAGGAAGGA | AGTGGATGGCACACGAAACA |
| SmTBCC | Samuk01G0035500 | 130 | AGCCTGACACCCTTTCTCTACC | CCAACCTCTTCTGCAGCCTT |
| SmUBC12 | Samuk11G0037500 | 102 | TTGTCTGGACCAACCAAAGGA | TGCTTCTTGCCAGGTGTTTTC |
| SmUBP | Samuk09G0089700 | 180 | TGCACAGTTGTTGCTCATGC | CTGCTTCATTTTCCCCGTGC |
表1 候选内参基因的引物
Table 1 Primers of candidate reference genes
| 内参基因 Reference genes | 基因ID Gene ID | 产物长度 Length of product /bp | 正向引物序列 Forward primer sequences(5'- 3') | 反向引物序列 Reverse primer sequences(5'- 3') |
|---|---|---|---|---|
| Sm18S | Samuk09G0136600 | 221 | AGATTGAAGGCGTTCTTTGGAT | TCAGTGTTTATGGACGGTGGAC |
| SmACT | Samuk13G0061200 | 129 | AGAAAGTTGGCCTCGCTGAA | CAGGAACCAGACCACCTGTC |
| SmEF-1α | Samuk14G0055700 | 101 | TCCAAGGCCAGGTACGATGA | AAACCAGAGATGGGGACGAAG |
| SmRPL1 | Samuk07G0011500 | 206 | CTGACCACCCCACCACTCTC | TCCTCCTCATCTGCCACCTC |
| SmRPS26 | Samuk08G0043900 | 230 | GCGAAGAAATGGAGGAAGGA | AGTGGATGGCACACGAAACA |
| SmTBCC | Samuk01G0035500 | 130 | AGCCTGACACCCTTTCTCTACC | CCAACCTCTTCTGCAGCCTT |
| SmUBC12 | Samuk11G0037500 | 102 | TTGTCTGGACCAACCAAAGGA | TGCTTCTTGCCAGGTGTTTTC |
| SmUBP | Samuk09G0089700 | 180 | TGCACAGTTGTTGCTCATGC | CTGCTTCATTTTCCCCGTGC |
| 内参基因 Reference gene | 基因ID Gene ID | 产物长度 Length of product /bp | 正向引物序列 Forward primer sequences(5'- 3') | 反向引物序列 Reverse primer sequence(5'- 3') |
|---|---|---|---|---|
| SmAACT4 | Samuk13G0079600 | 132 | CCTGTTTTGAGGGCATTGATTG | ACCAGCATAGAACGCAGCCA |
| SmDXS4 | Samuk14G0066700 | 194 | AACAATGCTCAGAAGCCGAGA | GCGATCCATCCAGTAATCCAC |
| SmFPS | Samuk04G0156300 | 131 | GAGGCAGAAGTAGGAACTCACCA | CCATTGTAGCAGTCAAGTGGTAAG |
| SmbAS1 | Samuk02G0323400 | 113 | GAGTGGGATACTGGTTTCGCTAT | TCCTTGACCTGAGATGCTTTGA |
| SmCYP716A-5 | Samuk08G0024600 | 146 | GCTGTTTTCTGTGGCCCTTC | CGAGTCTTGGCAATTTTCCC |
| SmUGT73C-14 | Samuk12G0007500 | 120 | ACCTACAAGTGCCCAAACAGAT | ATTCCCAGTTCACCACATTCC |
| SmbHLH8 | Samuk10G0068800 | 107 | CAACCTTTGACTCCGACTCCA | CCTTCCCTTACCCTAACCTCC |
| SmERF25 | Samuk11G0050000 | 129 | CAACAGCCACACCAACAGCA | TCTTCTCGGGTCACGGATCTC |
表2 目的基因的引物
Table 2 Primers of objective genes
| 内参基因 Reference gene | 基因ID Gene ID | 产物长度 Length of product /bp | 正向引物序列 Forward primer sequences(5'- 3') | 反向引物序列 Reverse primer sequence(5'- 3') |
|---|---|---|---|---|
| SmAACT4 | Samuk13G0079600 | 132 | CCTGTTTTGAGGGCATTGATTG | ACCAGCATAGAACGCAGCCA |
| SmDXS4 | Samuk14G0066700 | 194 | AACAATGCTCAGAAGCCGAGA | GCGATCCATCCAGTAATCCAC |
| SmFPS | Samuk04G0156300 | 131 | GAGGCAGAAGTAGGAACTCACCA | CCATTGTAGCAGTCAAGTGGTAAG |
| SmbAS1 | Samuk02G0323400 | 113 | GAGTGGGATACTGGTTTCGCTAT | TCCTTGACCTGAGATGCTTTGA |
| SmCYP716A-5 | Samuk08G0024600 | 146 | GCTGTTTTCTGTGGCCCTTC | CGAGTCTTGGCAATTTTCCC |
| SmUGT73C-14 | Samuk12G0007500 | 120 | ACCTACAAGTGCCCAAACAGAT | ATTCCCAGTTCACCACATTCC |
| SmbHLH8 | Samuk10G0068800 | 107 | CAACCTTTGACTCCGACTCCA | CCTTCCCTTACCCTAACCTCC |
| SmERF25 | Samuk11G0050000 | 129 | CAACAGCCACACCAACAGCA | TCTTCTCGGGTCACGGATCTC |
| 内参基因 Reference gene | 扩增效率 Amplification efficiency /% | 相关系数R2 Correlation coefficient R2 | 斜率K Slope K |
|---|---|---|---|
| Sm18S | 97.872 | 0.998 | -3.374 |
| SmACT | 93.727 | 0.999 | -3.482 |
| SmEF-1α | 99.789 | 0.999 | -3.327 |
| SmRPL1 | 100.206 | 0.999 | -3.317 |
| SmRPS26 | 94.469 | 0.999 | -3.462 |
| SmTBCC | 97.196 | 0.997 | -3.391 |
| SmUBC12 | 96.881 | 0.999 | -3.399 |
| SmUBP | 98.032 | 0.999 | -3.370 |
表3 候选内参基因扩增效率和标准曲线参数
Table 3 Amplification efficiency and standard curve para-meters of candidate reference genes
| 内参基因 Reference gene | 扩增效率 Amplification efficiency /% | 相关系数R2 Correlation coefficient R2 | 斜率K Slope K |
|---|---|---|---|
| Sm18S | 97.872 | 0.998 | -3.374 |
| SmACT | 93.727 | 0.999 | -3.482 |
| SmEF-1α | 99.789 | 0.999 | -3.327 |
| SmRPL1 | 100.206 | 0.999 | -3.317 |
| SmRPS26 | 94.469 | 0.999 | -3.462 |
| SmTBCC | 97.196 | 0.997 | -3.391 |
| SmUBC12 | 96.881 | 0.999 | -3.399 |
| SmUBP | 98.032 | 0.999 | -3.370 |

图2 候选内参基因的Ct值 箱须的上、下边分别表示Ct值中的最大值和最小值,箱体的上、下边分别表示Ct值中的上四分位数和下四分位数,箱体中的横线表示Ct值的中位数,实心菱形表示异常值,空心正方形表示平均值
Fig. 2 Ct values of candidate reference genes The upper and lower parts of the box whiskers indicate the maximum and minimum values of the Ct value;the upper and lower parts of the box body indicate the upper and lower quartiles of the Ct value;lines across the boxes depict the medians;solid diamonds represent outliers and hollow squares represent averages
| 内参基因 Reference gene | geNorm | NormFinder | BestKeeper | Delta Ct | RefFinder综合排序 Comprehensive ranking | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M值 M value | 排序 Rank | SV值 SV value | 排序 Rank | SD值 SD value | 排序 Rank | CV 值 CV value/% | 排序 Rank | r | 排序 Rank | 平均标准偏差 STDEV | 排序 Rank | 基因稳定性Gene stability | 排序 Rank | ||||||
| Sm18S | 0.710 | 6 | 0.722 | 7 | 1.41 | 1 | 5.71 | 4 | 0.950 | 5 | 0.900 | 7 | 4.300 | 5 | |||||
| SmACT | 0.373 | 1 | 0.347 | 1 | 1.51 | 4 | 6.04 | 5 | 0.985 | 1 | 0.670 | 1 | 1.410 | 1 | |||||
| SmEF-1α | 0.799 | 7 | 0.943 | 8 | 1.54 | 6 | 7.19 | 7 | 0.922 | 6 | 1.070 | 8 | 7.740 | 8 | |||||
| SmRPL1 | 0.373 | 1 | 0.373 | 3 | 1.53 | 5 | 5.65 | 3 | 0.985 | 1 | 0.680 | 2 | 2.450 | 2 | |||||
| SmRPS26 | 0.656 | 5 | 0.666 | 6 | 1.68 | 7 | 7.65 | 8 | 0.982 | 2 | 0.860 | 6 | 6.450 | 7 | |||||
| SmTBCC | 0.615 | 4 | 0.501 | 5 | 1.45 | 3 | 5.18 | 1 | 0.973 | 4 | 0.770 | 5 | 4.400 | 6 | |||||
| SmUBC12 | 0.508 | 2 | 0.480 | 4 | 1.53 | 5 | 6.31 | 6 | 0.979 | 3 | 0.740 | 4 | 3.940 | 4 | |||||
| SmUBP | 0.547 | 3 | 0.362 | 2 | 1.43 | 2 | 5.59 | 2 | 0.985 | 1 | 0.700 | 3 | 2.630 | 3 | |||||
表4 GeNorm,NormFinder和BestKeeper分析结果和排名
Table 4 GeNorm,NormFinder and BestKeeper analysis results and rankings
| 内参基因 Reference gene | geNorm | NormFinder | BestKeeper | Delta Ct | RefFinder综合排序 Comprehensive ranking | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M值 M value | 排序 Rank | SV值 SV value | 排序 Rank | SD值 SD value | 排序 Rank | CV 值 CV value/% | 排序 Rank | r | 排序 Rank | 平均标准偏差 STDEV | 排序 Rank | 基因稳定性Gene stability | 排序 Rank | ||||||
| Sm18S | 0.710 | 6 | 0.722 | 7 | 1.41 | 1 | 5.71 | 4 | 0.950 | 5 | 0.900 | 7 | 4.300 | 5 | |||||
| SmACT | 0.373 | 1 | 0.347 | 1 | 1.51 | 4 | 6.04 | 5 | 0.985 | 1 | 0.670 | 1 | 1.410 | 1 | |||||
| SmEF-1α | 0.799 | 7 | 0.943 | 8 | 1.54 | 6 | 7.19 | 7 | 0.922 | 6 | 1.070 | 8 | 7.740 | 8 | |||||
| SmRPL1 | 0.373 | 1 | 0.373 | 3 | 1.53 | 5 | 5.65 | 3 | 0.985 | 1 | 0.680 | 2 | 2.450 | 2 | |||||
| SmRPS26 | 0.656 | 5 | 0.666 | 6 | 1.68 | 7 | 7.65 | 8 | 0.982 | 2 | 0.860 | 6 | 6.450 | 7 | |||||
| SmTBCC | 0.615 | 4 | 0.501 | 5 | 1.45 | 3 | 5.18 | 1 | 0.973 | 4 | 0.770 | 5 | 4.400 | 6 | |||||
| SmUBC12 | 0.508 | 2 | 0.480 | 4 | 1.53 | 5 | 6.31 | 6 | 0.979 | 3 | 0.740 | 4 | 3.940 | 4 | |||||
| SmUBP | 0.547 | 3 | 0.362 | 2 | 1.43 | 2 | 5.59 | 2 | 0.985 | 1 | 0.700 | 3 | 2.630 | 3 | |||||
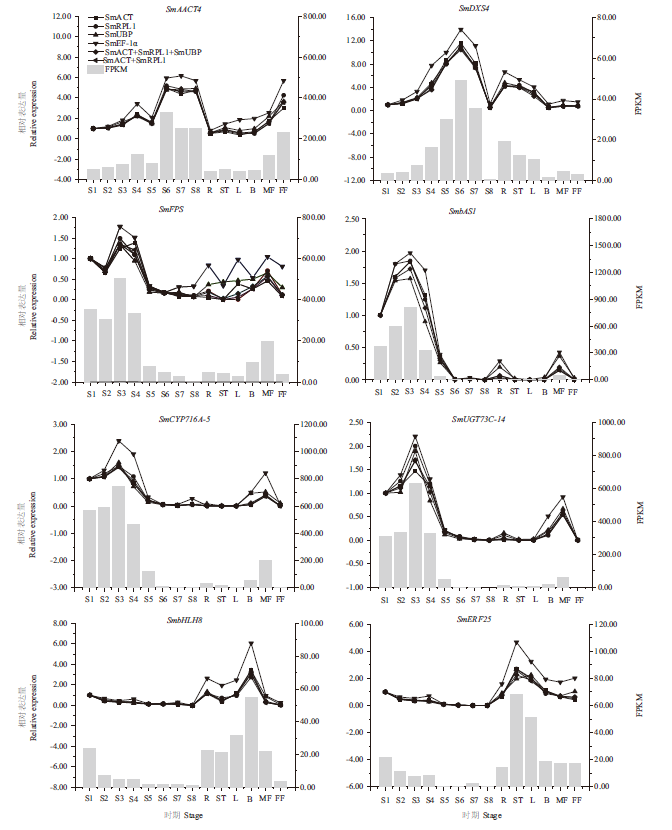
图4 无患子三萜皂苷生物合成途径中相关基因的RT-qPCR结果 R,根;ST,茎;L,叶;B,芽;MF,雄花;FF,雌花;S1,花后15 d的果皮;S2,花后45 d的果皮;S3,花后75 d的果皮;S4,花后90 d的果皮;S5,花后105 d的果皮;S6,花后120 d的果皮;S7,花后135 d的果皮;S8,花后150 d的果皮
Fig. 4 RT-qPCR expression pattern of genes related to the triterpenoid saponin biosynthesis of S. mukorossi R,root;ST,stem;L,leaf;B,bud;MF,male flower;FF,female flower;S1,pericarp at 15 d after flower pollination;S2,pericarp at 45 d after flower pollination;S3,pericarp at 75 d after flower pollination;S4,pericarp at 90 d after flower pollination;S5,pericarp at 105 d after flower pollination;S6,pericarp at 120 d after flower pollination;S7,pericarp at 135 d after flower pollination;S8,pericarp at 150 d after flower pollination
| [1] |
Kou XY, Zhang L, Yang SZ, et al. Selection and validation of reference genes for quantitative RT-PCR analysis in peach fruit under different experimental conditions[J]. Sci Hortic, 2017, 225:195-203.
doi: 10.1016/j.scienta.2017.07.004 URL |
| [2] |
Bustin SA, Benes V, Garson JA, et al. The MIQE guidelines:minimum information for publication of quantitative real-time PCR experiments[J]. Clin Chem, 2009, 55(4):611-622.
doi: 10.1373/clinchem.2008.112797 URL |
| [3] |
Nolan T, Hands RE, Bustin SA. Quantification of mRNA using real-time RT-PCR[J]. Nat Protoc, 2006, 1(3):1559-1582.
doi: 10.1038/nprot.2006.236 pmid: 17406449 |
| [4] |
Xu LF, Xu H, Cao YW, et al. Validation of reference genes for quantitative real-time PCR during bicolor tepal development in Asiatic hybrid lilies(Lilium spp. )[J]. Front Plant Sci, 2017, 8:669.
doi: 10.3389/fpls.2017.00669 URL |
| [5] |
Ferradás Y, Rey L, Martínez Ó, et al. Identification and validation of reference genes for accurate normalization of real-time quantitative PCR data in kiwifruit[J]. Plant Physiol Biochem, 2016, 102:27-36.
doi: 10.1016/j.plaphy.2016.02.011 URL |
| [6] |
Wu JY, Zhang HN, Liu LQ, et al. Validation of reference genes for RT-qPCR studies of gene expression in preharvest and postharvest longan fruits under different experimental conditions[J]. Front Plant Sci, 2016, 7:780.
doi: 10.3389/fpls.2016.00780 pmid: 27375640 |
| [7] |
袁伟, 万红建, 杨悦俭. 植物实时荧光定量PCR内参基因的特点及选择[J]. 植物学报, 2012, 47(4):427-436.
doi: 10.3724/SP.J.1259.2012.00427 |
| Yuan W, Wan HJ, Yang YJ. Characterization and selection of reference genes for real-time quantitative RT-PCR of plants[J]. Chin Bull Bot, 2012, 47(4):427-436. | |
| [8] | 王丽平, 梁瑾, 谌琴琴, 等. 千里光RT-qPCR分析中内参基因的选择[J]. 中国中药杂志, 2019, 44(3):465-471. |
| Wang LP, Liang J, Shen QQ, et al. Identification of stable reference gene by RT-qPCR in Senecio scandens[J]. China J Chin Mater Med, 2019, 44(3):465-471. | |
| [9] | 徐碧霞, 郭巧生, 朱再标, 等. 老鸦瓣实时定量PCR内参基因的筛选和验证[J]. 中国中药杂志, 2021, 46(4):938-943. |
| Xu BX, Guo QS, Zhu ZB, et al. Selection and validation of reference genes for quantitative Real-time PCR analysis in Amana edulis[J]. China J Chin Mater Med, 2021, 46(4):938-943. | |
| [10] |
Li DD, Hu B, Wang Q, et al. Identification and evaluation of reference genes for accurate transcription normalization in safflower under different experimental conditions[J]. PLoS One, 2015, 10(10):e0140218.
doi: 10.1371/journal.pone.0140218 URL |
| [11] |
Nicot N, Hausman JF, Hoffmann L, et al. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress[J]. J Exp Bot, 2005, 56(421):2907-2914.
pmid: 16188960 |
| [12] | 贾黎明, 孙操稳. 生物柴油树种无患子研究进展[J]. 中国农业大学学报, 2012, 17(6):191-196. |
| Jia LM, Sun CW. Research progress of biodiesel tree Sapindus mukorossi[J]. J China Agric Univ, 2012, 17(6):191-196. | |
| [13] | 徐圆圆, 周思维, 陈仲, 等. 无患子不同器官中的总皂苷和总黄酮含量[J]. 南京林业大学学报:自然科学版, 2021, 45(4):83-89. |
| Xu YY, Zhou SW, Chen Z, et al. Contents of the total saponins and total flavonoids in different organs of Sapindus mukorossi[J]. J Nanjing For Univ Nat Sci Ed, 2021, 45(4):83-89. | |
| [14] | 刘济铭, 陈仲, 孙操稳, 等. 无患子属种质资源种实性状变异及综合评价[J]. 林业科学, 2019, 55(6):44-54. |
| Liu JM, Chen Z, Sun CW, et al. Variation in fruit and seed properties and comprehensive assessment of germplasm resources of the genus Sapindus[J]. Sci Silvae Sin, 2019, 55(6):44-54. | |
| [15] | 徐圆圆, 贾黎明, 陈仲, 等. 无患子三萜皂苷研究进展[J]. 化学通报, 2018, 81(12):1078-1088. |
| Xu YY, Jia LM, Chen Z, et al. Advances on triterpenoid saponin of Sapindus mukorossi[J]. Chemistry, 2018, 81(12):1078-1088. | |
| [16] |
Sun CW, Wang LC, Liu JM, et al. Genetic structure and biogeographic divergence among Sapindus species:an inter-simple sequence repeat-based study of germplasms in China[J]. Ind Crops Prod, 2018, 118:1-10.
doi: 10.1016/j.indcrop.2018.03.029 URL |
| [17] |
Zhao GC, Gao YH, Gao SL, et al. The phenological growth stages of Sapindus mukorossi according to BBCH scale[J]. Forests, 2019, 10(6):462.
doi: 10.3390/f10060462 URL |
| [18] |
Gao Y, Gao SL, Jia LM, et al. Canopy characteristics and light distribution in Sapindus mukorossi Gaertn. are influenced by crown architecture manipulation in the hilly terrain of Southeast China[J]. Sci Hortic, 2018, 240:11-22.
doi: 10.1016/j.scienta.2018.05.034 URL |
| [19] |
Hu QW, Chen YY, Jiao QY, et al. Triterpenoid saponins from the pulp of Sapindus mukorossi and their antifungal activities[J]. Phytochemistry, 2018, 147:1-8.
doi: 10.1016/j.phytochem.2017.12.004 URL |
| [20] | 陈莎莎, 陈丽萍, 林继辉. 无患子果皮皂苷提取工艺研究[J]. 云南民族大学学报:自然科学版, 2019, 28(6):558-562. |
| Chen SS, Chen LP, Lin JH. The extraction technology of saponins from the soapberry pericarp[J]. J Yunnan Minzu Univ Nat Sci Ed, 2019, 28(6):558-562. | |
| [21] |
Xu YY, Gao Y, Chen Z, et al. Metabolomics analysis of the soapberry(Sapindus mukorossi Gaertn.)pericarp during fruit development and ripening based on UHPLC-HRMS[J]. Sci Rep, 2021, 11(1):11657.
doi: 10.1038/s41598-021-91143-0 URL |
| [22] | Vandesompele J, de Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes[J]. Genome Biol, 2002, 3(7):RESEARCH0034. |
| [23] |
Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data:a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets[J]. Cancer Res, 2004, 64(15):5245-5250.
doi: 10.1158/0008-5472.CAN-04-0496 URL |
| [24] |
Pfaffl MW, Tichopad A, Prgomet C, et al. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity:BestKeeper—Excel-based tool using pair-wise correlations[J]. Biotechnol Lett, 2004, 26(6):509-515.
doi: 10.1023/B:BILE.0000019559.84305.47 URL |
| [25] |
Xie F, Xiao P, Chen D, et al. miRDeepFinder:a miRNA analysis tool for deep sequencing of plant small RNAs[J]. Plant Mol Biol, 2012. DOI: 10.1007/s11103-012-9885-2.
doi: 10.1007/s11103-012-9885-2 |
| [26] | 蒋婷婷, 高燕会, 童再康. 石蒜属植物实时荧光定量PCR内参基因的选择[J]. 园艺学报, 2015, 42(6):1129-1138. |
| Jiang TT, Gao YH, Tong ZK. Selection of reference genes for quantitative real-time PCR in Lycoris[J]. Acta Hortic Sin, 2015, 42(6):1129-1138. | |
| [27] |
Dudziak K, Sozoniuk M, Szczerba H, et al. Identification of stable reference genes for qPCR studies in common wheat(Triticum aestivum L.)seedlings under short-term drought stress[J]. Plant Methods, 2020, 16:58.
doi: 10.1186/s13007-020-00601-9 URL |
| [28] | 曹映辉, 郑燕, 张燕萍, 等. 建兰花香物质合成相关基因RT-qPCR内参基因筛选[J]. 分子植物育种, 2021, http://kns.cnki.net/kcms/detail/46.1068.S.20210510.1555.008.html. |
| Cao YH, Zheng Y, Zhang YP, et al. Selection of suitable RT-qPCR reference genes for floral scent biosynthesis in Cymbidium ensifolium[J]. Mol Plant Breed, 2021, http://kns.cnki.net/kcms/detail/46.1068.S.20210510.1555.008.html. | |
| [29] | 章颖佳, 程少禹, 王卓为, 等. 紫玉兰‘红元宝’花芽分化阶段基因定量分析的内参基因筛选[J]. 广西植物, 2022, 42(1):113-121. |
| Zhang YJ, Cheng SY, Wang ZW, et al. Selection of reference genes in Magnolia liliflora ‘Hongyuanbao’ during flower bud differentiation[J]. Guihaia, 2022, 42(1):113-121. | |
| [30] |
de Spiegelaere W, Dern-Wieloch J, Weigel R, et al. Reference gene validation for RT-qPCR, a note on different available software packages[J]. PLoS One, 2015, 10(3):e0122515.
doi: 10.1371/journal.pone.0122515 URL |
| [31] | 张玉芳, 赵丽娟, 曾幼玲. 基因表达研究中内参基因的选择与应用[J]. 植物生理学报, 2014, 50(8):1119-1125. |
| Zhang YF, Zhao LJ, Zeng YL. Selection and application of reference genes for gene expression studies[J]. Plant Physiol J, 2014, 50(8):1119-1125. | |
| [32] |
Hwang HS, Lee H, Choi YE. Transcriptomic analysis of Siberian ginseng(Eleutherococcus senticosus)to discover genes involved in saponin biosynthesis[J]. BMC Genomics, 2015, 16(1):180.
doi: 10.1186/s12864-015-1357-z URL |
| [33] |
Han JY, Chun JH, Oh SA, et al. Transcriptomic analysis of Kalopanax septemlobus and characterization of KsBAS, CYP716A94 and CYP72A397 genes involved in hederagenin saponin biosynthesis[J]. Plant Cell Physiol, 2018, 59(2):319-330.
doi: 10.1093/pcp/pcx188 URL |
| [34] |
Shan CM, Wang CK, Zhang SX, et al. Transcriptome analysis of Clinopodium gracile(Benth.)Matsum and identification of genes related to Triterpenoid Saponin biosynthesis[J]. BMC Genomics, 2020, 21(1):49.
doi: 10.1186/s12864-020-6454-y URL |
| [35] | Xiao Z, Sun XB, Liu XQ, et al. Selection of reliable reference genes for gene expression studies on Rhododendron molle G. don[J]. Front Plant Sci, 2016, 7:1547. |
| 徐圆圆, 陈仲, 贾黎明, 等. 植物三萜皂苷生物合成途径及调控机制研究进展[J]. 中国科学:生命科学, 2021, 51(5):525-555. | |
|
Xu YY, Chen Z, Jia LM, et al. Advances in understanding of the biosynthetic pathway and regulatory mechanism of triterpenoid saponins in plants[J]. Sci Sin Vitae, 2021, 51(5):525-555.
doi: 10.1360/SSV-2020-0230 URL |
|
| [37] |
Zhao YJ, Li C. Biosynthesis of plant triterpenoid saponins in microbial cell factories[J]. J Agric Food Chem, 2018, 66(46):12155-12165.
doi: 10.1021/acs.jafc.8b04657 URL |
| [38] |
Augustin JM, Kuzina V, Andersen SB, et al. Molecular activities, biosynthesis and evolution of triterpenoid saponins[J]. Phytochemistry, 2011, 72(6):435-457.
doi: 10.1016/j.phytochem.2011.01.015 pmid: 21333312 |
| [39] |
Xu YY, Zhao GC, Ji XQ, et al. Metabolome and transcriptome analysis reveals the transcriptional regulatory mechanism of triterpenoid saponin biosynthesis in soapberry(Sapindus mukorossi Gaertn.)[J]. J Agric Food Chem, 2022, DOI: 10.1021/acs.jafc.2c01672.
doi: 10.1021/acs.jafc.2c01672 |
| [1] | 王佳蕊, 孙培媛, 柯瑾, 冉彬, 李洪有. 苦荞糖基转移酶基因FtUGT143的克隆及表达分析[J]. 生物技术通报, 2023, 39(8): 204-212. |
| [2] | 孙明慧, 吴琼, 刘丹丹, 焦小雨, 王文杰. 茶树CsTMFs的克隆与表达分析[J]. 生物技术通报, 2023, 39(7): 151-159. |
| [3] | 余慧, 王静, 梁昕昕, 辛亚平, 周军, 赵会君. 宁夏枸杞铁镉响应基因的筛选及其功能验证[J]. 生物技术通报, 2023, 39(7): 195-205. |
| [4] | 赵雪婷, 高利燕, 王俊刚, 沈庆庆, 张树珍, 李富生. 甘蔗AP2/ERF转录因子基因ShERF3的克隆、表达及其编码蛋白的定位[J]. 生物技术通报, 2023, 39(6): 208-216. |
| [5] | 姜晴春, 杜洁, 王嘉诚, 余知和, 王允, 柳忠玉. 虎杖转录因子PcMYB2的表达特性和功能分析[J]. 生物技术通报, 2023, 39(5): 217-223. |
| [6] | 姚姿婷, 曹雪颖, 肖雪, 李瑞芳, 韦小妹, 邹承武, 朱桂宁. 火龙果溃疡病菌实时荧光定量PCR内参基因的筛选[J]. 生物技术通报, 2023, 39(5): 92-102. |
| [7] | 王艺清, 王涛, 韦朝领, 戴浩民, 曹士先, 孙威江, 曾雯. 茶树SMAS基因家族的鉴定及互作分析[J]. 生物技术通报, 2023, 39(4): 246-258. |
| [8] | 宋海娜, 吴心桐, 杨鲁豫, 耿喜宁, 张华敏, 宋小龙. 葱鳞葡萄孢菌诱导下韭菜RT-qPCR内参基因的筛选和验证[J]. 生物技术通报, 2023, 39(3): 101-115. |
| [9] | 刘思佳, 王浩楠, 付宇辰, 闫文欣, 胡增辉, 冷平生. ‘西伯利亚’百合LiCMK基因克隆及功能分析[J]. 生物技术通报, 2023, 39(3): 196-205. |
| [10] | 王涛, 漆思雨, 韦朝领, 王艺清, 戴浩民, 周喆, 曹士先, 曾雯, 孙威江. CsPPR和CsCPN60-like在茶树白化叶片中的表达分析及互作蛋白验证[J]. 生物技术通报, 2023, 39(3): 218-231. |
| [11] | 庞强强, 孙晓东, 周曼, 蔡兴来, 张文, 王亚强. 菜心BrHsfA3基因克隆及其对高温胁迫的响应[J]. 生物技术通报, 2023, 39(2): 107-115. |
| [12] | 穆德添, 万凌云, 章瑶, 韦树根, 陆英, 付金娥, 田艺, 潘丽梅, 唐其. 钩藤管家基因筛选及生物碱合成相关基因的表达分析[J]. 生物技术通报, 2023, 39(2): 126-138. |
| [13] | 苗淑楠, 高宇, 李昕儒, 蔡桂萍, 张飞, 薛金爱, 季春丽, 李润植. 大豆GmPDAT1参与油脂合成和非生物胁迫应答的功能分析[J]. 生物技术通报, 2023, 39(2): 96-106. |
| [14] | 葛雯冬, 王腾辉, 马天意, 范震宇, 王玉书. 结球甘蓝PRX基因家族全基因组鉴定与逆境条件下的表达分析[J]. 生物技术通报, 2023, 39(11): 252-260. |
| [15] | 杨旭妍, 赵爽, 马天意, 白玉, 王玉书. 三个甘蓝WRKY基因的克隆及其对非生物胁迫的表达[J]. 生物技术通报, 2023, 39(11): 261-269. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||