








生物技术通报 ›› 2026, Vol. 42 ›› Issue (4): 101-113.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0928
• 研究报告 •
苏燕竹( ), 李达, 张爱爱, 刘永光, 张秀荣(
), 李达, 张爱爱, 刘永光, 张秀荣( ), 薛其勤(
), 薛其勤( )
)
收稿日期:2025-08-27
出版日期:2026-02-09
发布日期:2026-02-09
通讯作者:
张秀荣,女,博士,副教授,研究方向 :作物与蔬菜种质资源创新;E-mail: zhangxiurong@wfust.edu.cn作者简介:苏燕竹,女,博士,讲师,研究方向 :大豆遗传育种及耐逆性;E-mail: suyanz1526@163.com
基金资助:
SU Yan-zhu( ), LI Da, ZHANG Ai-ai, LIU Yong-guang, ZHANG Xiu-rong(
), LI Da, ZHANG Ai-ai, LIU Yong-guang, ZHANG Xiu-rong( ), XUE Qi-qin(
), XUE Qi-qin( )
)
Received:2025-08-27
Published:2026-02-09
Online:2026-02-09
摘要:
目的 研究大豆肉桂醇脱氢酶(Cinnamyl Alcohol Dehydrogenase, CAD)基因家族(GmCADs)成员特征和在不同胁迫下的表达模式,为后续深入解析GmCADs基因家族生物学功能奠定基础。 方法 基于大豆基因组数据,利用生物信息学方法对GmCADs基因家族成员进行鉴定,对其编码蛋白质的特征、系统发育关系、基因结构、保守基序等进行分析。 结果 在大豆全基因组水平鉴定了18个CAD家族基因(GmCAD1-GmCAD18),分布在13条染色体上,氨基酸数量在219-364 aa之间。系统发育分析显示,GmCADs基因家族分为4个亚族,同一亚族基因具有相似的基因结构和保守基序。共线性分析表明片段复制是GmCADs基因家族扩张的主要形式。选择压力分析表明,GmCADs基因处于纯化选择。GmCADs基因家族的启动子上含有丰富的与大豆光响应、激素响应、逆境响应和生长发育过程相关的顺式作用元件。通过5个间接蛋白,所有GmCADs基因形成了一个复杂的蛋白质相互作用网络,且GO功能分析显著富集在CAD活性和木质素代谢过程等条目。GmCADs成员在不同大豆组织和逆境下(干旱、盐、冷、荫蔽和高温)表达具有一定的选择性,其中GmCAD11和GmCAD18表达量较高。RT-qPCR验证表明,在盐和干旱处理后,9个GmCADs基因表达量在不同时间显著改变,主要表现为上调表达。 结论 18个大豆CAD成员在分布、结构及功能上存在多样性,且GmCADs基因可能在大豆植株生长发育过程中响应非生物胁迫。
苏燕竹, 李达, 张爱爱, 刘永光, 张秀荣, 薛其勤. 大豆CAD基因家族的鉴定及表达分析[J]. 生物技术通报, 2026, 42(4): 101-113.
SU Yan-zhu, LI Da, ZHANG Ai-ai, LIU Yong-guang, ZHANG Xiu-rong, XUE Qi-qin. Identification and Expression Analysis of CAD Gene Family in Soybean(Glycine max (L.) Merr.)[J]. Biotechnology Bulletin, 2026, 42(4): 101-113.
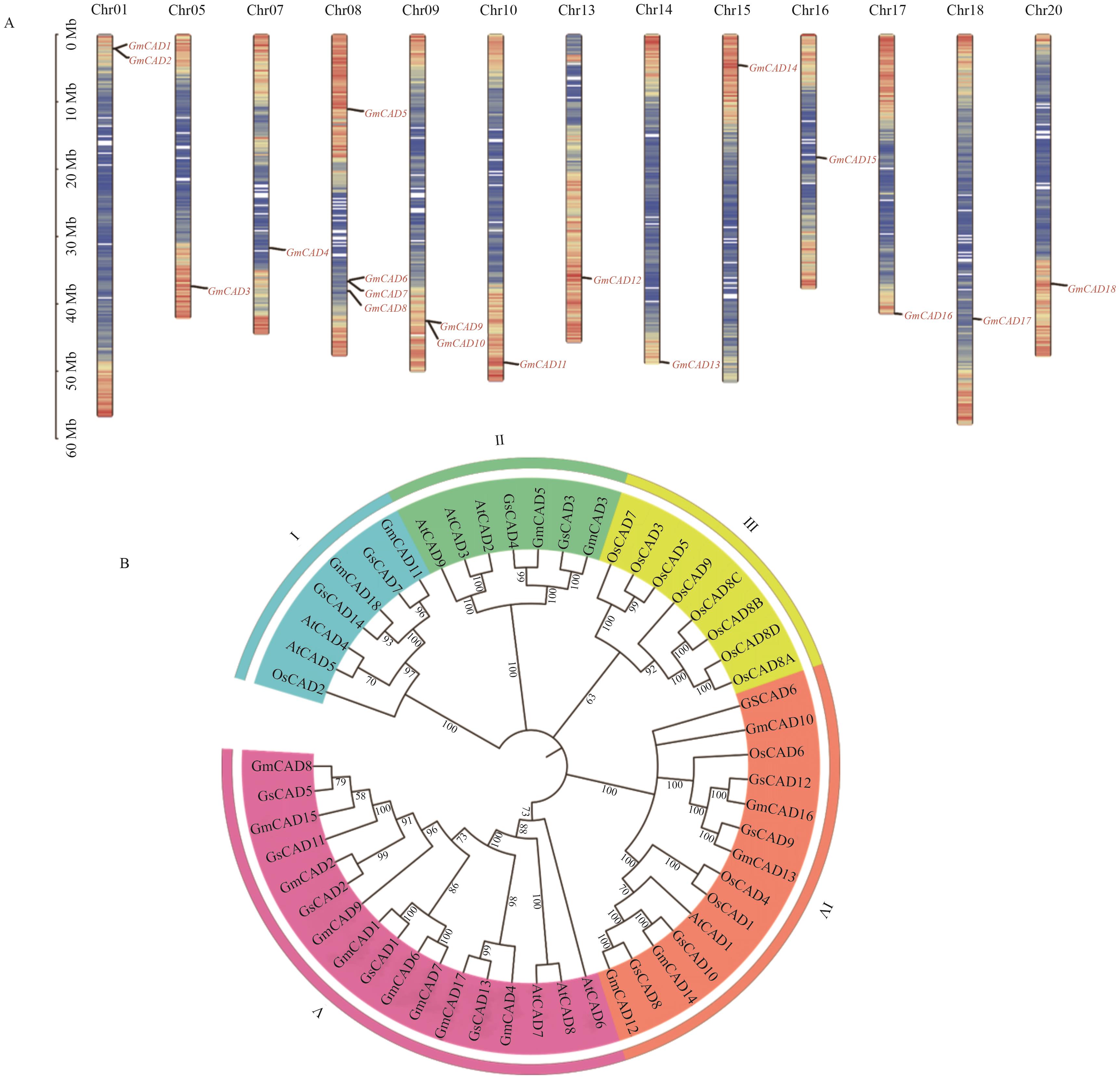
图1 GmCADs基因在大豆染色体上的分布(A)和系统进化分析(B)A:染色体上的颜色表示基因的密度,颜色越红,密度越高,颜色越蓝,密度越低
Fig. 1 Chromosomal distribution (A) and phylogenetic analysis (B) of GmCAD genesA: The colors on the chromosome indicate the density of the gene. The redder the color, the higher the density. The bluer the color, the lower the density
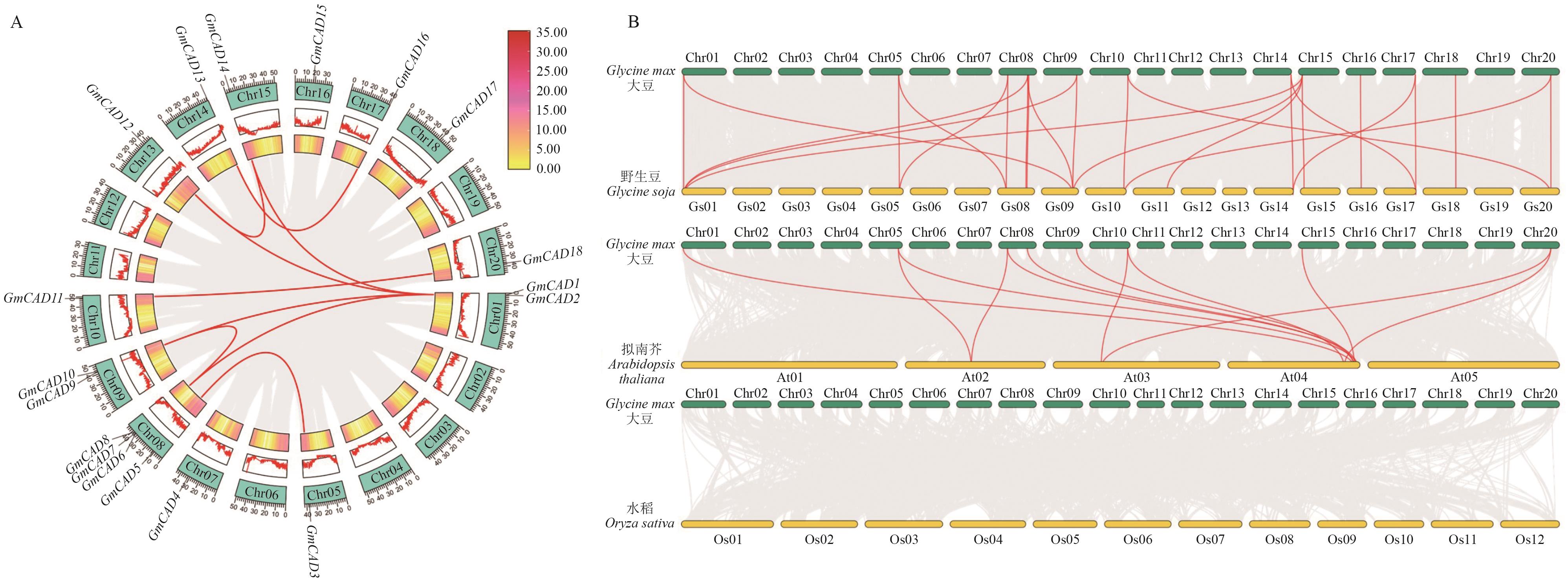
图3 CAD基因家族共线性分析A:大豆CAD家族成员种内共线性分析。红色曲线连接片段复制的基因对;外侧部分指示基因在染色体上的对应位置,绿色框对应不同的染色体(Chr01-Chr20);红色锯齿线突出显示了每条染色体上的基因密度高低;图的内侧框进一步强调了染色体内的基因密度,图例表示基因密度,黄色到红色表示基因密度由小变大。B:大豆与不同物种间CAD基因的共线性分析。红色高亮线为GmCADs基因的物种间共线性对,灰色线条为大豆与其他物种基因组的所有共线性区块
Fig. 3 Colinear analysis in the CAD gene familyA: Intraspecific colinear analysis of GmCAD genes family. Red curved line connects duplicated gene pairs. The outer part indicates the locations of these genes on the chromosome, and the green boxs correspond to different chromosomes (Chr 01-Chr 20). Red serrated lines highlight the gene density on each chromosome. The inner frames of the chart further emphasize the gene density in the chromosome, and with yellow to red indicating a change from small to large gene density. B: Colinear analysis of CAD gene between soybean and other species. The red lines highlight collinear pairs of GmCAD genes, while the gray lines in the background indicate collinear blocks between the genome of soybean and the other species
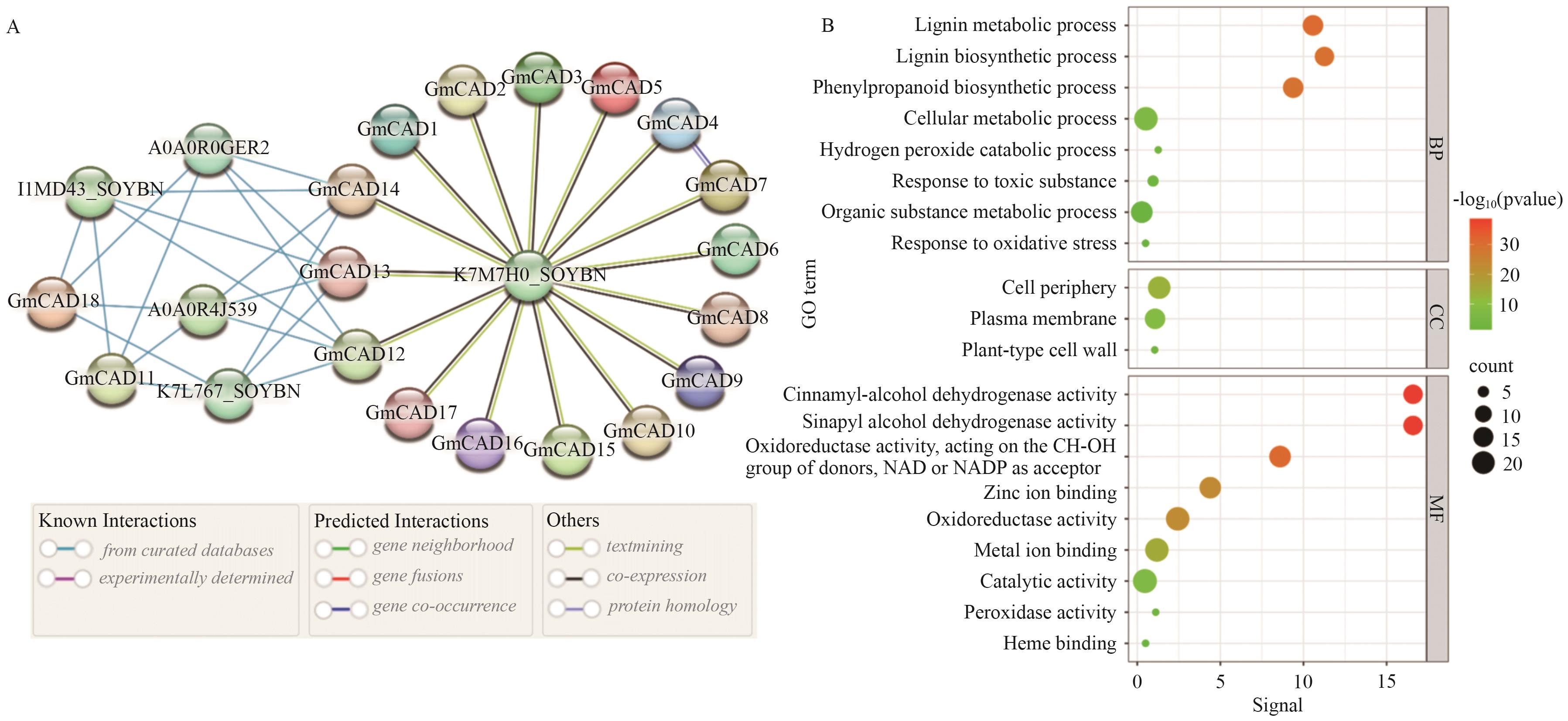
图5 GmCADs基因的蛋白质相互作用网络(A)和GO富集分析(B)BP:生物过程;CC:细胞组分;MF:分子功能
Fig. 5 Protein-protein interaction (PPI) network (A) and GO enrichment analysis (B) of GmCAD genesBP: Biological process. CC: Cellular component. MF: Molecular function
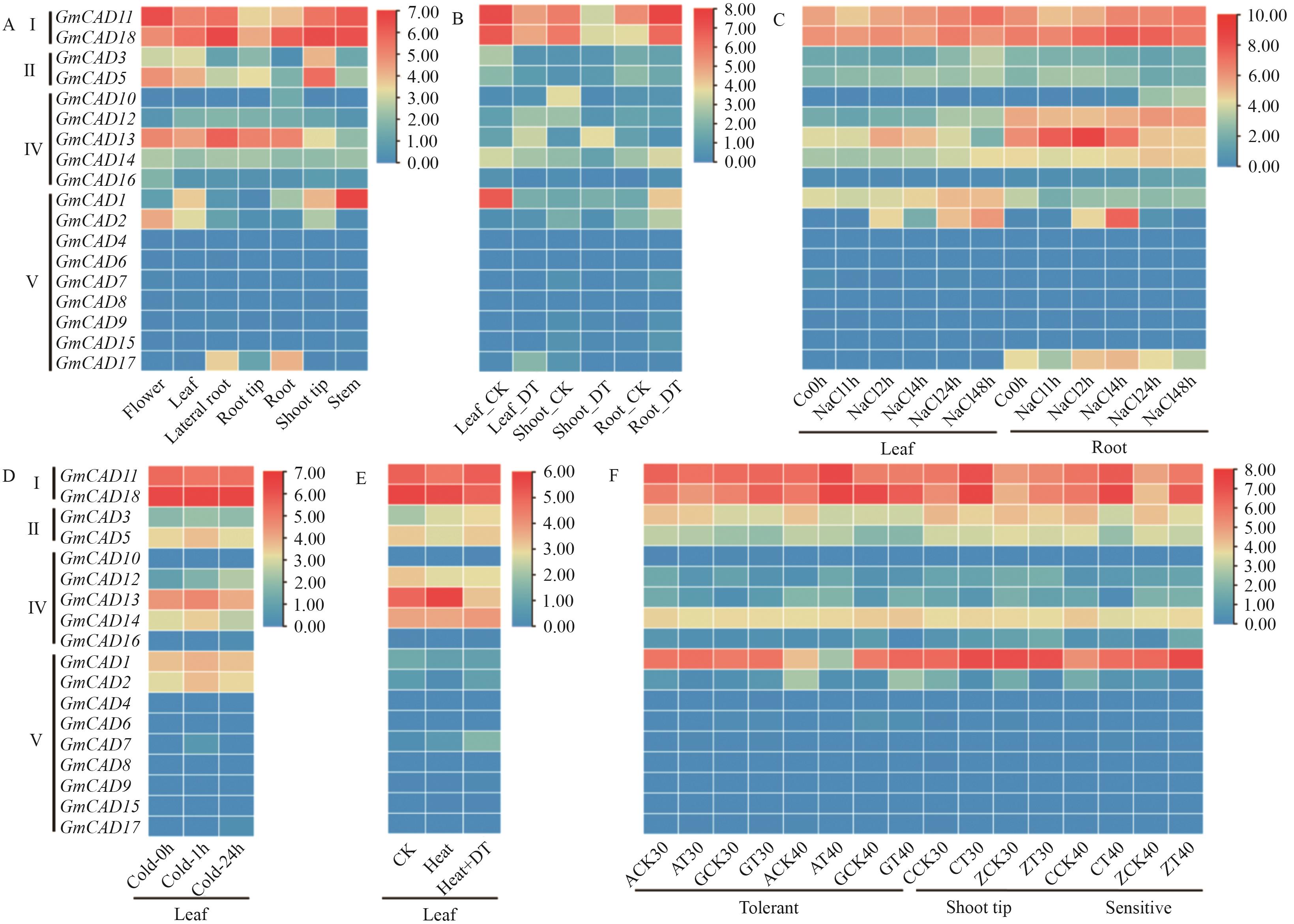
图6 大豆GmCADs家族基因组织表达特性(A)和不同胁迫条件下表达模式分析(B-F)B-F:分别是干旱、荫蔽、寒冷、高温和盐胁迫下GmCADs基因的表达模式。图中CK均表示对照,DT表示干旱胁迫,图C中“A”是耐荫大豆,“C”是不耐荫大豆,“T”是遮荫处理
Fig. 6 Tissue-specific expression characteristics (A) and expression patterns analysis under different stress conditions (B-F) of GmCAD family genes in soybean(Glycine max (L.) Merr.)B-F: the expression patterns of the GmCAD genes under conditions of drought (DT), shade, cold, heat and salt stress, respectively. In the figures, CK refers to the control, and DT to drought stress. In Figure C, ‘A’ refers to shade-tolerant soybeans, ‘C’ to shade-intolerant soybeans, and ‘T’ to shading treatment
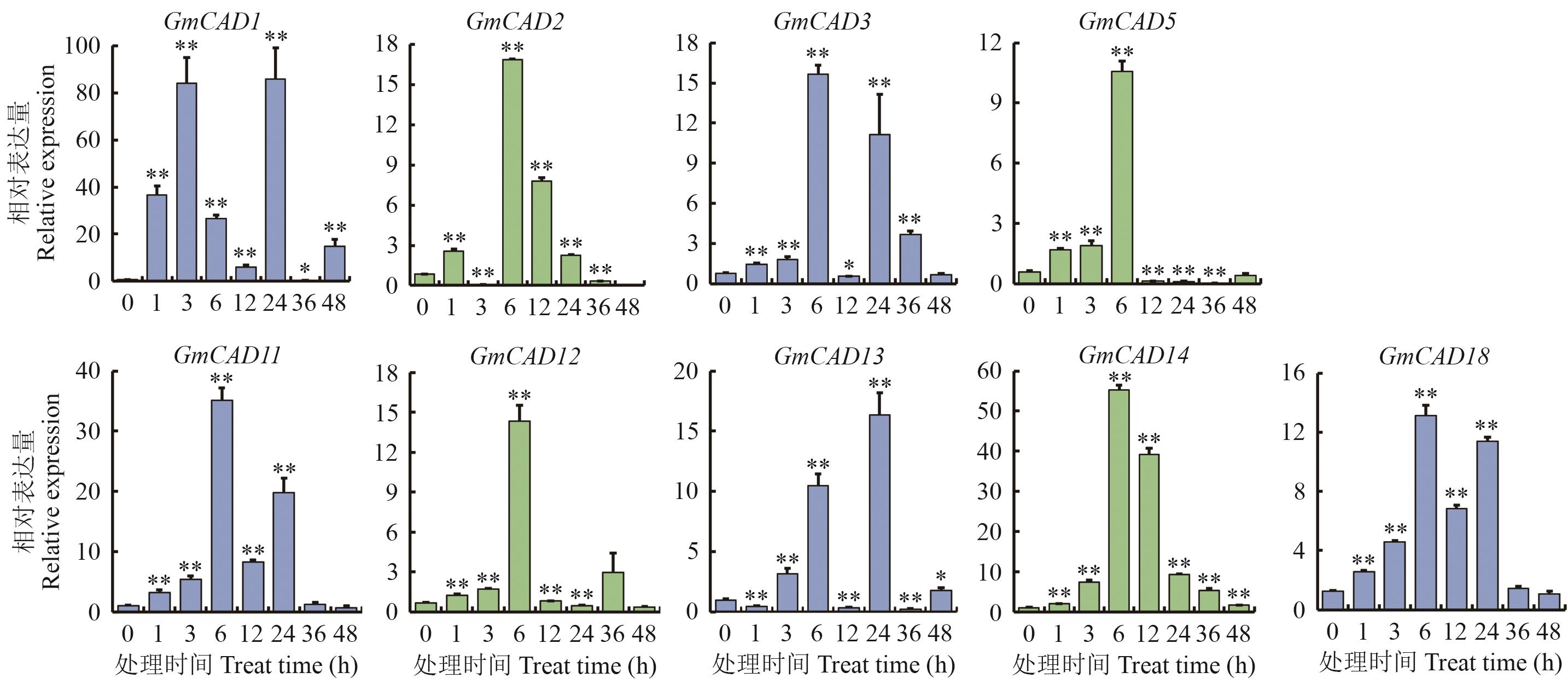
图7 盐胁迫下CAD基因的表达量*和**分别表示盐胁迫处理后1-48 h与处理前0 h比较表达量具有显著(P<0.05)和极显著差异(P<0.01),下同
Fig. 7 The CAD genes expression under salt stress*and ** indicate that there are significant difference and extremely significant differences in expressions when comparing 1 h to 48 h after salt stress treatment with 0 before the treatment, with P<0.05 and P<0.01, the same below
| [1] | 刘欣婷, 王娟, 侯献飞, 等. 木质素及其合成基因在作物抗倒伏中的功能及其研究进展 [J]. 分子植物育种, 2019, 17(2): 655-662. |
| Liu XT, Wang J, Hou XF, et al. Progress of functions of lignin and relevant genes in plant lodging resistance [J]. Mol Plant Breed, 2019, 17(2): 655-662. | |
| [2] | 刘威. 薄皮甜瓜肉桂醇脱氢酶(CAD)在非生物胁迫下参与木质素合成的功能探究 [D]. 沈阳: 沈阳农业大学, 2019. |
| Liu W. Functional research of CmCADs in lignin biosynthesis in oriental melon (Cucumis melo L.) under abiotic stresses [D]. Shenyang: Shenyang Agricultural University, 2019. | |
| [3] | Gross GG, Stöckigt J, Mansell RL, et al. Three novel enzymes involved in the reduction of ferulic acid to coniferyl alcohol in higher plants: ferulate: Co a ligase, feruloyl-Co a reductase and coniferyl alcohol oxidoreductase [J]. FEBS Lett, 1973, 31(3): 283-286. |
| [4] | Knight ME, Halpin C, Schuch W. Identification and characterisation of cDNA clones encoding cinnamyl alcohol dehydrogenase from tobacco [J]. Plant Mol Biol, 1992, 19(5): 793-801. |
| [5] | Kim SJ, Kim MR, Bedgar DL, et al. Functional reclassification of the putative cinnamyl alcohol dehydrogenase multigene family in Arabidopsis [J]. Proc Natl Acad Sci U S A, 2004, 101(6): 1455-1460. |
| [6] | Tobias CM, Chow EK. Structure of the cinnamyl-alcohol dehydrogenase gene family in rice and promoter activity of a member associated with lignification [J]. Planta, 2005, 220(5): 678-688. |
| [7] | Saballos A, Ejeta G, Sanchez E, et al. A genomewide analysis of the cinnamyl alcohol dehydrogenase family in sorghum [sorghum bicolor (L.) moench] identifies SbCAD2 as the brown midrib6 gene [J]. Genetics, 2009, 181(2): 783-795. |
| [8] | 任晓庆, 王波, 欧阳春平, 等. 谷子CAD基因家族的鉴定及分析 [J]. 山西农业科学, 2022, 50(3): 289-295. |
| Ren XQ, Wang B, Ouyang CP, et al. Identification and analysis of millet CAD gene family [J]. J Shanxi Agric Sci, 2022, 50(3): 289-295. | |
| [9] | An FF, Chen T, Zhu WL, et al. Systematic analysis of cinnamyl alcohol dehydrogenase family in cassava and validation of MeCAD13 and MeCAD28 in lignin synthesis and postharvest physiological deterioration [J]. Int J Mol Sci, 2024, 25(21): 11668. |
| [10] | Lu SS, Liu W, Yang LJ, et al. Transcriptome and VcCADs gene family analyses reveal mechanisms of blight resistance in rabbiteye blueberry [J]. Front Plant Sci, 2025, 16: 1601658. |
| [11] | Park HL, Kim TL, Bhoo SH, et al. Biochemical characterization of the rice cinnamyl alcohol dehydrogenase gene family [J]. Molecules, 2018, 23(10): 2659. |
| [12] | Fan L, Shi WJ, Hu WR, et al. Molecular and biochemical evidence for phenylpropanoid synthesis and presence of wall-linked phenolics in cotton fibers [J]. J Integr Plant Biol, 2009, 51(7): 626-637. |
| [13] | Guillaumie S, Pichon M, Martinant JP, et al. Differential expression of phenylpropanoid and related genes in brown-midrib bm1, bm2, bm3, and bm4 young near-isogenic maize plants [J]. Planta, 2007, 226(1): 235-250. |
| [14] | Rong W, Luo M, Shan T, et al. A wheat cinnamyl alcohol dehydrogenase TaCAD12 contributes to host resistance to the sharp eyespot disease [J]. Front Plant Sci, 2016, 7: 1723. |
| [15] | 刘堰珺, 马静, 王广龙, 等. 胡萝卜肉桂醇脱氢酶基因的克隆及其对非生物胁迫的响应 [J]. 西北植物学报, 2016, 36(7): 1294-1301. |
| Liu YJ, Ma J, Wang GL, et al. Cloning and expression profile analysis of the gene encoding cinnamyl alcohol dehydrogenase under abiotic stress in carrot [J]. Acta Bot Boreali Occidentalia Sin, 2016, 36(7): 1294-1301. | |
| [16] | Liu W, Jin YZ, Li MM, et al. Analysis of CmCADs and three lignifying enzymes in oriental melon (‘CaiHong7’) seedlings in response to three abiotic stresses [J]. Sci Hortic, 2018, 237: 257-268. |
| [17] | Liu W, Jiang Y, Wang CH, et al. Lignin synthesized by CmCAD2 and CmCAD3 in oriental melon (Cucumis melo L.) seedlings contributes to drought tolerance [J]. Plant Mol Biol, 2020, 103(6): 689-704. |
| [18] | Raza A, Asghar MA, Javed HH, et al. Optimum nitrogen improved stem breaking resistance of intercropped soybean by modifying the stem anatomical structure and lignin metabolism [J]. Plant Physiol Biochem, 2023, 199: 107720. |
| [19] | Gu Y, Zheng HY, Li S, et al. Effects of narrow-wide row planting patterns on canopy photosynthetic characteristics, bending resistance and yield of soybean in maize-soybean intercropping systems [J]. Sci Rep, 2024, 14: 9361. |
| [20] | 孙洪吉. 大豆GmCAD1基因的抗旱功能研究 [D]. 长春: 吉林农业大学, 2024. |
| Sun HJ. Study on the drought resistance function of soybean GmCAD1 gene [D]. Changchun: Jilin Agricultural University, 2024. | |
| [21] | 金贺. 大豆胞囊线虫胁迫下微紫青霉处理对GmC4H和GmCAD的影响及抗性机制研究 [D]. 沈阳: 沈阳农业大学, 2022. |
| Jin H. Effects of Penicillium purpureus on GmC4H and GmCAD under soybean cyst nematode stress and their resistance mechanism [D]. Shenyang: Shenyang Agricultural University, 2022. | |
| [22] | Chen CJ, Wu Y, Li JW, et al. TBtools-II: a “one for all, all for one” bioinformatics platform for biological big-data mining [J]. Mol Plant, 2023, 16(11): 1733-1742. |
| [23] | Kumar S, Stecher G, Li M, et al. MEGA X: molecular evolutionary genetics analysis across computing platforms [J]. Mol Biol Evol, 2018, 35(6): 1547-1549. |
| [24] | Bailey TL, Boden M, Buske FA, et al. MEME Suite: tools for motif discovery and searching [J]. Nucleic Acids Res, 2009, 37(): W202-W208. |
| [25] | Hu B, Jin JP, Guo AY, et al. GSDS 2.0: an upgraded gene feature visualization server [J]. Bioinformatics, 2015, 31(8): 1296-1297. |
| [26] | Lescot M, Déhais P, Thijs G, et al. PlantCARE, a database of plant Cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences [J]. Nucleic Acids Res, 2002, 30(1): 325-327. |
| [27] | Almeida-Silva F, Pedrosa-Silva F, Venancio TM. The Soybean Expression Atlas v2: a comprehensive database of over 5000 RNA-seq samples [J]. Plant J, 2023, 116(4): 1041-1051. |
| [28] | Su YZ, Hao XS, Zeng WY, et al. Genome-wide association with transcriptomics reveals a shade-tolerance gene network in soybean [J]. Crop J, 2024, 12(1): 232-243. |
| [29] | Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method [J]. Methods, 2001, 25(4): 402-408. |
| [30] | 胡文冉, 郝晓燕, 赵准, 等. 棉花CAD基因家族的生物信息学分析 [J]. 生物技术进展, 2023, 13(3): 412-424. |
| Hu WR, Hao XY, Zhao Z, et al. Bioinformatic analysis of the CAD gene family in cotton [J]. Curr Biotechnol, 2023, 13(3): 412-424. | |
| [31] | Cannon SB, Mitra A, Baumgarten A, et al. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana . [J]. BMC Plant Biol, 2004, 4(1): 10. |
| [32] | Sibout R, Eudes A, Mouille G, et al. CINNAMYL ALCOHOL DEHYDROGENASE-C and -d are the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis [J]. Plant Cell, 2005, 17(7): 2059-2076. |
| [33] | Hirano K, Aya K, Kondo M, et al. OsCAD2 is the major CAD gene responsible for monolignol biosynthesis in rice culm [J]. Plant Cell Rep, 2012, 31(1): 91-101. |
| [34] | 尹言言, 刘靖, 郑炳松, 等. 脱落酸调控植物非生物胁迫研究进展 [J]. 核农学报, 2025, 39(9): 1916-1927. |
| Yin YY, Liu J, Zheng BS, et al. Research progress of abscisic acid in regulating plant abiotic stress [J]. J Nucl Agric Sci, 2025, 39(9): 1916-1927. | |
| [35] | Cheng H, Li LL, Xu F, et al. Expression patterns of a cinnamyl alcohol dehydrogenase gene involved in lignin biosynthesis and environmental stress in Ginkgo biloba [J]. Mol Biol Rep, 2013, 40(1): 707-721. |
| [36] | Deng WW, Zhang M, Wu JQ, et al. Molecular cloning, functional analysis of three cinnamyl alcohol dehydrogenase (CAD) genes in the leaves of tea plant, Camellia sinensis [J]. J Plant Physiol, 2013, 170(3): 272-282. |
| [37] | Kim YH, Bae JM, Huh GH. Transcriptional regulation of the cinnamyl alcohol dehydrogenase gene from sweet potato in response to plant developmental stage and environmental stress [J]. Plant Cell Rep, 2010, 29(7): 779-791. |
| [38] | 龙国辉, 武鹏雨, 付嘉智, 等. 过氧化物酶调控木质素合成研究进展 [J]. 现代农业科技, 2021(23): 47-49, 54. |
| Long GH, Wu PY, Fu JZ, et al. Research progress on regulation of peroxidase on lignin synthesis [J]. Mod Agric Sci Technol, 2021(23): 47-49, 54. | |
| [39] | Barceló AR, Ros LVG, Carrasco AE. Looking for syringyl peroxidases [J]. Trends Plant Sci, 2007, 12(11): 486-491. |
| [1] | 贺启琛, 杨扬, 阿丽亚·外力, 唐新月, 李忠喜, 陈永坤, 陈凌娜. 薰衣草CuAO基因家族特征及LaCuAO1降解生物胺功能研究[J]. 生物技术通报, 2026, 42(1): 114-124. |
| [2] | 杨丹, 靳雅荣, 毛春力, 王碧娴, 张雅宁, 杨智怡, 周芷瑶, 杨锐鸣, 范恒睿, 黄琳凯, 严海东. 象草C2H2基因家族鉴定及表达分析[J]. 生物技术通报, 2026, 42(1): 251-261. |
| [3] | 淦晨露, 游雨婷, 谢菡萏, 曾子贤, 朱博. 植物黄素单加氧酶研究进展[J]. 生物技术通报, 2026, 42(1): 1-12. |
| [4] | 任云儿, 伍国强, 成斌, 魏明. 甜菜BvATGs基因家族全基因组鉴定及盐胁迫下表达模式分析[J]. 生物技术通报, 2026, 42(1): 184-197. |
| [5] | 杨跃琴, 邢英, 仲子荷, 田维军, 杨雪清, 王建旭. 甲基汞胁迫下水稻OsMATE34的表达及功能分析[J]. 生物技术通报, 2026, 42(1): 86-94. |
| [6] | 李健斌, 侯家娥, 李雷林, 艾明涛, 刘天泰, 崔秀明, 杨千. 三七脂氧合酶的全基因组鉴定及其对茉莉酸甲酯和创伤的响应[J]. 生物技术通报, 2026, 42(1): 218-229. |
| [7] | 张月, 戴月华, 张莹莹, 李奥辉, 李楚慧, 薛金爱, 秦慧彬, 陈妍, 聂萌恩, 张海平. 大豆烯酰辅酶A还原酶ECR14基因的克隆与功能分析[J]. 生物技术通报, 2026, 42(1): 95-104. |
| [8] | 陈静欢, 房国楠, 朱文豪, 叶广继, 苏旺, 贺苗苗, 杨生龙, 周云. 马铃薯种质资源淀粉表征及相关基因表达分析[J]. 生物技术通报, 2026, 42(1): 170-183. |
| [9] | 龙林茜, 曾银萍, 王茜, 邓玉萍, 葛敏茜, 陈彦灼, 李鑫娟, 杨军, 邹建. 向日葵GH3基因家族鉴定及其在花发育中的功能分析[J]. 生物技术通报, 2026, 42(1): 125-138. |
| [10] | 李亚涛, 张志鹏, 赵梦瑶, 吕镇, 甘恬, 魏浩, 吴书凤, 马玉超. 根瘤菌Bd1的全基因组分析及TetR3对细胞生长和结瘤的负调控功能[J]. 生物技术通报, 2025, 41(9): 289-301. |
| [11] | 李珊, 马登辉, 马红义, 姚文孔, 尹晓. 葡萄SKP1基因家族鉴定与表达分析[J]. 生物技术通报, 2025, 41(9): 147-158. |
| [12] | 黄国栋, 邓宇星, 程宏伟, 但焱南, 周会汶, 吴兰花. 大豆ZIP基因家族鉴定及响应铝胁迫的表达分析[J]. 生物技术通报, 2025, 41(9): 71-81. |
| [13] | 巩慧玲, 邢玉洁, 马俊贤, 蔡霞, 冯再平. 马铃薯LAC基因家族的鉴定及盐胁迫下表达分析[J]. 生物技术通报, 2025, 41(9): 82-93. |
| [14] | 关陟昊, 单治易, 熊赫, 赵瑞雪. 基于计算文献的大豆耦合性状知识发现研究[J]. 生物技术通报, 2025, 41(9): 345-356. |
| [15] | 程婷婷, 刘俊, 王利丽, 练从龙, 魏文君, 郭辉, 吴尧琳, 杨晶凡, 兰金旭, 陈随清. 杜仲查尔酮异构酶基因家族全基因组鉴定及其表达模式分析[J]. 生物技术通报, 2025, 41(9): 242-255. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||