








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (6): 225-235.doi: 10.13560/j.cnki.biotech.bull.1985.2020-1389
Previous Articles Next Articles
Received:2020-11-15
Online:2021-06-26
Published:2021-07-08
WU Feng-zhang, WANG He-xin. Low Temperature Stress Response Mediated by Protein Ubiquitination in Plant[J]. Biotechnology Bulletin, 2021, 37(6): 225-235.
| 类型 Type | 泛素连接酶 Ub-ligase | 靶目标 Target(s) | 物种 Species | 一般性功能描述 General function description | 参考文献 References |
|---|---|---|---|---|---|
| RING | HOS1 | ICE1 | 拟南芥 Arabidopsis thaliana | 负调控抗冻性 Negatively regulate freezing resistance | [22] |
| MaSINA1 | MaICE1 | 香蕉 Musa acuminata | 负调控耐冷性 Negatively regulate chilling tolerance | [32] | |
| MdMIEL1 | MdMYB308L | 苹果 Malus domestica | 负调控抗冻性 Negatively regulate freezing resistance | [31] | |
| SAP9 | Rad23d | 拟南芥 Arabidopsis thaliana | 正调控抗寒性 Positively regulates cold resistance | [61] | |
| U-box | CaPUB1 | RPN6 | 水稻 Oryza sativa | 正调控耐冷性 Positively regulates chilling tolerance | [56] |
| PUB25/26 | MYB15 | 拟南芥 Arabidopsis thaliana | 正调控抗冻性 Positively regulates freezing resistance | [29] | |
| AtCHIP | PP2A | 拟南芥 Arabidopsis thaliana | 负调控耐冷性 Negatively regulate chilling tolerance | [60] | |
| SCF | EBF1/2 | PIF3 | 拟南芥 Arabidopsis thaliana | 正调控抗冻性 Positively regulates freezing resistance | [30] |
| EBF1/2 | EIN3/EIL1 | 拟南芥 Arabidopsis thaliana | 正调控抗冻性 Positively regulates freezing resistance | [49-50] | |
| FBP7 | LOS1? | 拟南芥 Arabidopsis thaliana | 负调控耐冷性 Negatively regulate chilling tolerance | [55] | |
| COI1 | JAZs | 拟南芥 Arabidopsis thaliana | 正调控抗冻性 Positively regulates freezing resistance | [51,53] | |
| DDB | COP1 | HY5 | 拟南芥 Arabidopsis thaliana | 负调控抗冻性 Negatively regulate freezing resistance | [34] |
| BTB | ETO1/EOL1/2 | ACS5 | 拟南芥 Arabidopsis thaliana | 负调控抗冻性 Negatively regulate freezing resistance | [45] |
Table 1 General functional descriptions of the E3 ligases involved low temperature stress responses in plant
| 类型 Type | 泛素连接酶 Ub-ligase | 靶目标 Target(s) | 物种 Species | 一般性功能描述 General function description | 参考文献 References |
|---|---|---|---|---|---|
| RING | HOS1 | ICE1 | 拟南芥 Arabidopsis thaliana | 负调控抗冻性 Negatively regulate freezing resistance | [22] |
| MaSINA1 | MaICE1 | 香蕉 Musa acuminata | 负调控耐冷性 Negatively regulate chilling tolerance | [32] | |
| MdMIEL1 | MdMYB308L | 苹果 Malus domestica | 负调控抗冻性 Negatively regulate freezing resistance | [31] | |
| SAP9 | Rad23d | 拟南芥 Arabidopsis thaliana | 正调控抗寒性 Positively regulates cold resistance | [61] | |
| U-box | CaPUB1 | RPN6 | 水稻 Oryza sativa | 正调控耐冷性 Positively regulates chilling tolerance | [56] |
| PUB25/26 | MYB15 | 拟南芥 Arabidopsis thaliana | 正调控抗冻性 Positively regulates freezing resistance | [29] | |
| AtCHIP | PP2A | 拟南芥 Arabidopsis thaliana | 负调控耐冷性 Negatively regulate chilling tolerance | [60] | |
| SCF | EBF1/2 | PIF3 | 拟南芥 Arabidopsis thaliana | 正调控抗冻性 Positively regulates freezing resistance | [30] |
| EBF1/2 | EIN3/EIL1 | 拟南芥 Arabidopsis thaliana | 正调控抗冻性 Positively regulates freezing resistance | [49-50] | |
| FBP7 | LOS1? | 拟南芥 Arabidopsis thaliana | 负调控耐冷性 Negatively regulate chilling tolerance | [55] | |
| COI1 | JAZs | 拟南芥 Arabidopsis thaliana | 正调控抗冻性 Positively regulates freezing resistance | [51,53] | |
| DDB | COP1 | HY5 | 拟南芥 Arabidopsis thaliana | 负调控抗冻性 Negatively regulate freezing resistance | [34] |
| BTB | ETO1/EOL1/2 | ACS5 | 拟南芥 Arabidopsis thaliana | 负调控抗冻性 Negatively regulate freezing resistance | [45] |
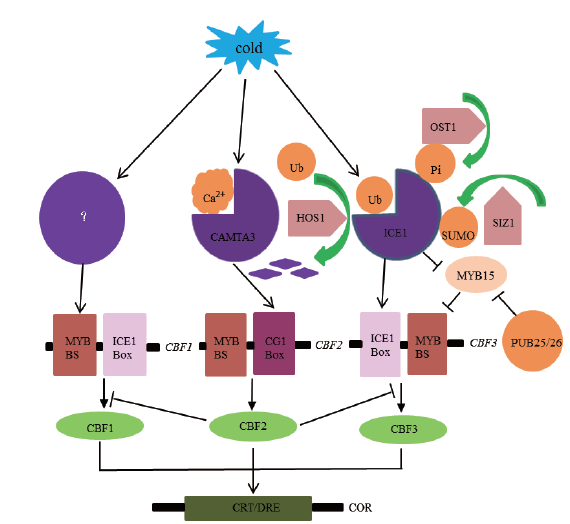
Fig. 1 Protein ubiquitination regulates the CBF-dependent low temperature signaling pathway CBFs transcription factors are responsible for the regulation of COR genes containing CRT/DRE(CCGAC)motifs in their promoters. CBFs are activated by ICE1 and CAMTA transcription factors,whereas repressed by MYB15. HOS1 and SIZ1 encode RING E3 ligase and SUMO E3 ligase,respectively,which antagonistically regulate the abundance of ICE1 protein. OST1 is a kinase that positively regulates the abundance of the ICE1 protein. Crooked arrows indicate post-translational regulation,and solid arrows indicate activation. T-shaped bars represent repression;the solid line represent direct interaction. The same below
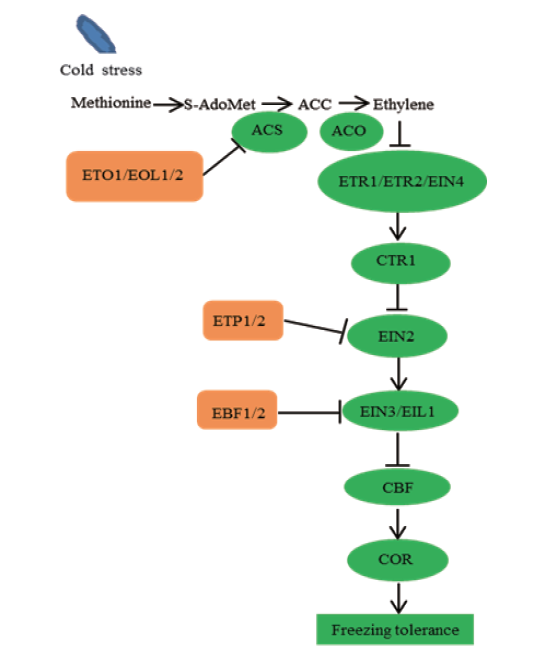
Fig. 2 Schematic diagram depicts the involvement of E3s in regulation of freezing stress tolerance by the ethylene(ET)pathway in Arabidopsis In the biosynthesis of ethylene,the ETO1,EOL1 and EOL2 modulate ET biosynthesis by interacting with the ET biosynthetic enzyme ACS5 and inhibiting its activity by proteasome-dependent degradation. In the signal transductions of ethylene,in the absence of ethylene,the highly phosphorylated ethylene receptors activate CTR1 kinase activity,which in turn phosphorylates EIN2,likely causing the degradation of EIN2 by F-box proteins EPT1 and EPT2. Meanwhile,EIN3/EILs are also subjected to proteasomal degradation mediated by F-box proteins EBF1 and EBF2. In the presence of ethylene,ethylene binding inactivates the receptors by suppressing its phosphorylation,which consequently leads to deactivation of CTR1. The un-phosphorylated EIN2 is thus cleaved and its C-terminal domain is translocated into the nucleus,resulting in activation of EIN3/EILs and downstream transcriptional cascades. Finally,they regulate the freezing stress tolerance response
| [1] |
Gong Z, Xiong L, Shi H, et al. Plant abiotic stress response and nutrient use efficiency[J]. Science China Life Sciences, 2020, 63(5):635-674.
doi: 10.1007/s11427-020-1683-x URL |
| [2] |
Liu J, Shi Y, Yang S. Insights into the regulation of CBF cold signaling in plants[J]. Journal of Integrative Plant Biology, 2018, 60(9):780-795.
doi: 10.1111/jipb.12657 URL |
| [3] |
Pearce RS. Plant freezing and damage[J]. Annals of Botany, 2001, 87(4):417-424.
doi: 10.1006/anbo.2000.1352 URL |
| [4] |
Steponkus PL. Role of the plasma membrane in freezing injury and cold acclimation[J]. Annual Review Plant Physiology and Molecular Biology, 1984, 35(1):543-584.
doi: 10.1146/annurev.pp.35.060184.002551 URL |
| [5] |
Thomashow MF. Plant cold acclimation:freezing tolerante genes and regulatory mechanisms[J]. Annual Review Plant Physiology and Molecular Biology, 1999, 50:571-599.
doi: 10.1146/annurev.arplant.50.1.571 URL |
| [6] |
Chinnusamy V, Zhu J, Zhu JK. Cold stress regulation of gene expression in plants[J]. Trends in Plant Science, 2007, 12(10):444-451.
doi: 10.1016/j.tplants.2007.07.002 URL |
| [7] |
Xiong LM, Schumaker KS, Zhu JK. Cell signaling during cold, drough, and salt stress[J]. The Plant Cell, 2002, 14:165-183.
doi: 10.1105/tpc.010278 URL |
| [8] |
Winfield MO, Lu C, Wilson ID, et al. Plant responses to cold:transcriptome analysis of wheat[J]. Plant Biotechnology Journal, 2010, 8(7):749-771.
doi: 10.1111/pbi.2010.8.issue-7 URL |
| [9] |
Thomashow MF. Molecular basis of plant cold acclimation:insights gained from studying the CBF cold response pathway[J]. Plant Physiology, 2010, 154(2):571-577.
doi: 10.1104/pp.110.161794 URL |
| [10] |
Catalá R, Salinas J. Regulatory mechanisms involved in cold acclimation response[J]. Spanish Journal of Agricultural Research, 2008, 6:211-220.
doi: 10.5424/sjar/200806S1-390 URL |
| [11] |
Nakasone MA, Livnat-Levanon N, Glickman MH, et al. Mixed-linkage ubiquitin chains send mixed messages[J]. Structure, 2013, 21(5):727-740.
doi: 10.1016/j.str.2013.02.019 pmid: 23562397 |
| [12] |
Komander D, Rape M. The ubiquitin code[J]. Annu Rev Biochem, 2012, 81(1):203-229.
doi: 10.1146/annurev-biochem-060310-170328 URL |
| [13] |
Thrower JS, Hoffman L, Rechsteiner M, et al. Recognition of the polyubiquitin proteolytic signal[J]. The EMBO Journal, 2000, 19(1):94-102.
doi: 10.1093/emboj/19.1.94 URL |
| [14] | Chen Q, Yang X, Xie Q. Approaches to determine protein ubiquitination residue types[M]// Lois LM, Matthiesen R. Plant Proteostasis:Methods and Protocols, Methods in Molecular Biology, New York: Humana Press, 2016:3-10. |
| [15] | Isono E, Nagel MK. Deubiquitylating enzymes and their emerging role in plant biology[J]. Frontiers in Plant Science, 2014, 5:56-62. |
| [16] |
Vierstra RD. The expanding universe of ubiquitin and ubiquitinlike modifiers[J]. Plant Physiology, 2012, 160(1):2-14.
doi: 10.1104/pp.112.200667 URL |
| [17] | Morreale FE, Walden H. SnapShot:Types of ubiquitin ligases[J]. Cell, 2016, 165:48-248. |
| [18] |
Vierstra RD. The ubiquitin-26S proteasome system at the nexus of plant biology[J]. Nature Reviews Molecular Cell Biology, 2009, 10(6):385-397.
doi: 10.1038/nrm2688 URL |
| [19] |
Biedermann S, Hellmann H. WD40 and CUL4-based E3 ligases:lubricating all of aspects life[J]. Trends in Plant Science, 2011, 16(11):38-46.
doi: 10.1016/j.tplants.2010.09.007 URL |
| [20] | 王丰, 施一公. 26S蛋白酶体的结构生物学研究进展[J]. 中国科学:生命科学, 2014, 44:965-974. |
|
Wang F, Shi YG. Progress in structural biology of 26S proteasome[J]. Scientia Sinica Vitae, 2014, 44:965-974.
doi: 10.1360/052014-162 URL |
|
| [21] |
Jia Y, Ding Y, Shi Y, et al. The cbfs triple mutants reveal the essential functions of CBFs in cold acclimation and allow the definition of CBF regulons in Arabidopsis[J]. New Phytologist, 2016, 212(2):345-353.
doi: 10.1111/nph.2016.212.issue-2 URL |
| [22] | Dong CH, Agarwal M, Zhang Y, et al. The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1[J]. Proceedings of the National Academy of Sciences of the United States of America, 2006, 103(21):8281-8286. |
| [23] |
Ding Y, Li H, Zhang X, et al. OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis[J]. Developmental Cell, 2015, 32(9):278-289.
doi: 10.1016/j.devcel.2014.12.023 URL |
| [24] |
Miura K, Ohta M, Nakazawa M, et al. ICE1 Ser403 is necessary for protein stabilization and regulation of cold signaling and tolerance[J]. The Plant Journal, 2011, 67(2):269-279.
doi: 10.1111/tpj.2011.67.issue-2 URL |
| [25] |
Miura K, Jin JB, Lee J, et al. SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis[J]. The Plant Cell, 2007, 19:1403-1414.
doi: 10.1105/tpc.106.048397 URL |
| [26] |
Zhan X, Zhu JK, Lang Z. Increasing freezing tolerance:kinase regulation of ICE1[J]. Developmental Cell, 2015, 32(3):257-258.
doi: 10.1016/j.devcel.2015.01.004 URL |
| [27] | Ye K, Li H, Ding Y, et al. BRASSINOSTEROID-INSENSITIVE2 negatively regulates the stability of transcription factor ICE1 in response to cold stress in Arabidopsis[J]. The Plant Cell, 2019, 31(9):2682-2696. |
| [28] |
Liu Z, Jia Y, Ding Y, et al. Plasma membrane CRPK1-mediated phosphorylation of 14-3-3 proteins induces their nuclear import to fine-tune CBF signaling during cold response[J]. Molecular Cell, 2017, 66(1):117-128.
doi: 10.1016/j.molcel.2017.02.016 URL |
| [29] |
Wang X, Ding Y, Li Z, et al. PUB25 and PUB26 promote plant freezing tolerance by degrading the cold signaling negative regulator MYB15[J]. Developmental Cell, 2019, 51(2):222-235.
doi: 10.1016/j.devcel.2019.08.008 URL |
| [30] | Jiang B, Shi Y, Zhang X, et al. PIF3 is a negative regulator of the CBF pathway and freezing tolerance in Arabidopsis[J]. Proceedings of the National Academy of Sciences of the United States of America, 2017, 114(32):6695-6702. |
| [31] |
An JP, Wang XF, Zhang XW, et al. An apple MYB transcription factor regulates cold tolerance and anthocyanin accumulation and undergoes MIEL1-mediated degradation[J]. Plant Biotechnology Journal, 2020, 18(2):337-353.
doi: 10.1111/pbi.v18.2 URL |
| [32] |
Fan ZQ, Chen JY, Kuang JF, et al. The banana fruit SINA ubiquitin ligase MaSINA1 regulates the stability of MaICE1 to be negatively involved in cold stress response[J]. Frontiers in Plant Science, 2017, 8:995-1007.
doi: 10.3389/fpls.2017.00995 URL |
| [33] |
Lau OS, Deng X. Plant hormone signaling lightens up:integrators of light and hormones[J]. Current Opinion in Plant Biology, 2010, 13(5):571-577.
doi: 10.1016/j.pbi.2010.07.001 URL |
| [34] | Catalá R, Medina J, Salinas J. Integration of low temperature and light signaling during cold acclimation response in Arabidopsis[J]. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108(39):16475-16480. |
| [35] |
Catalá R, López-Cobollo R, Mar Castellano M, et al. The Arabidopsis 14-3-3 protein RARE COLD INDUCIBLE 1A links low-temperature response and ethylene biosynjournal to regulate freezing tolerance and cold acclimation[J]. The Plant Cell, 2014, 26:3326-3342.
doi: 10.1105/tpc.114.127605 URL |
| [36] |
Zhao M, Liu W, Xia X, et al. Cold acclimation-induced freezing tolerance of Medicago truncatula seedlings is negatively regulated by ethylene[J]. Physiologia Plantarum, 2014, 152(1):115-129.
doi: 10.1111/ppl.12161 URL |
| [37] |
Zhao D, Shen L, Fan B, et al. Ethylene and cold participate in the regulation of LeCBF1 gene expression in postharvest tomato fruits[J]. FEBS Letters, 2009, 583(20):3329-3334.
doi: 10.1016/j.febslet.2009.09.029 URL |
| [38] |
Zhang Z, Huang R. Enhanced tolerance to freezing in tobacco and tomato Overexpressing transcription factor TERF2/LeERF2 is modulated by ethylene biosynjournal[J]. Plant Molecular Biology, 2010, 73(3):241-249.
doi: 10.1007/s11103-010-9609-4 URL |
| [39] |
Munshaw GC, Ervin EH, Beasley JS, et al. Effects of late-season ethephon applications on cold tolerance parameters of four bermudagrass cultivars[J]. Crop Science, 2010, 50(3):1022-1029.
doi: 10.2135/cropsci2008.09.0565 URL |
| [40] |
Shi YT, Tian SW, Hou LY, et al. Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis[J]. The Plant Cell, 2012, 24(6):2578-2595.
doi: 10.1105/tpc.112.098640 URL |
| [41] |
Janowiak F, Dorffling K. Chilling-induced changes in the contents of 1-aminocyclopropane-1-carboxylic acid(ACC)and its N-malonyl conjugate(MACC)in seedlings of two maize inbreds differing in chilling tolerance[J]. Journal of Plant Physiology, 1995, 147(2):257-262.
doi: 10.1016/S0176-1617(11)81514-2 URL |
| [42] |
Yang SF, Hoffman NE. Ethylene biosynjournal and its regulation in higher-plants[J]. Annual Review of Plant Physiology, 1984, 35(1):155-189.
doi: 10.1146/annurev.pp.35.060184.001103 URL |
| [43] |
Gingerich DJ, Gagne JM, Salter DW, et al. Cullins 3a and 3b assemble with members of the broad complex/tramtrack/bric-a-brac(BTB)protein family to form essential ubiquitinprotein ligases(E3s)in Arabidopsis[J]. The Journal of Biological Chemistry, 2005, 280(4):18810-18821.
doi: 10.1074/jbc.M413247200 URL |
| [44] |
Wang KL, Yoshida H, Lurin C, et al. Regulation of ethylene gas biosynjournal by the Arabidopsis ETO1 protein[J]. Nature, 2004, 428:945-950.
doi: 10.1038/nature02516 URL |
| [45] |
Christians MJ, Gingerich DJ, Hansen M, et al. The BTB ubiquitin ligases ETO1, EOL1 and EOL2 act collectively to regulate ethylene biosynjournal in Arabidopsis by controlling type-2 ACC synthase levels[J]. The Plant Journal, 2009, 57(2):332-345.
doi: 10.1111/j.1365-313X.2008.03693.x pmid: 18808454 |
| [46] | Catalá R, Salinas J. The Arabidopsis ethylene overproducer mutant eto1-3 displays enhanced freezing tolerance[J]. Plant Signaling & Behavior, 2015, 10(3):e989768. |
| [47] | Ju C, Yoon GM, Shemansky JM, et al. CTR1 phosphorylates the central regulator EIN2 to control ethylene mone signaling from hebrane to the nucleus in Arabidopsis[J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(47):9486-19491. |
| [48] |
Qiao H, Chang KN, Yazaki J, et al. Interplay between ethylene, ETP1/ETP2 F-box proteins, and degradation of EIN2 triggers thylene responses in Arabidopsis[J]. Genes & Development, 2009, 23:512-521.
doi: 10.1101/gad.1765709 URL |
| [49] |
Wen X, Zhang C, Ji Y, et al. Activation of ethylene signaling is mediated by nuclear translocation of the cleaved EIN2 carboxyl terminus[J]. Cell Research, 2012, 22(11):1613-1616.
doi: 10.1038/cr.2012.145 URL |
| [50] |
Potuschak T, Lechner E, Parmentier Y, et al. EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F-box proteins:EBF1 and EBF2[J]. Cell, 2003, 115(6):679-689.
pmid: 14675533 |
| [51] |
Hu Y, Jiang L, Wang F, et al. Jasmonate regulates the inducer of cbf expression- C-repeat binding factor/DRE binding factor1 cascade and freezing tolerance in Arobidopsis[J]. The Plant Cell, 2013, 25:2907-2924.
doi: 10.1105/tpc.113.112631 URL |
| [52] | Du H, Liu HB, Xiong LZ. Endogenous auxin and jasmonic acid levels differentially modulated by abiotic stresses in rice[J]. Frontiers in Plant Science, 2013, 4:389-397. |
| [53] |
Kazan K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance[J]. Trends in Plant Science, 2015, 20(4):219-229.
doi: 10.1016/j.tplants.2015.02.001 URL |
| [54] |
Calderón-Villalobos LA, Nill C, Marrocco K, et al. The evolutionarily conserved Arabidopsis thaliana F-box protein AtFBP7 is required for efficient translation during temperature stress[J]. Gen, 2007, 392(1/2):106-116.
doi: 10.1016/j.gene.2006.11.016 URL |
| [55] | Guo Y, Xiong L, Ishitani M, et al. An Arabidopsis mutation in translation elongation factor 2 causes superinduction of CBF/DREB1 transcription factor genes but blocks the induction of their downstream targets under low temperatures[J]. Proceedings of the National Academy of Sciences of the United States of America, 2002, 99(11):7786-7791. |
| [56] |
Cho K SK, Chung HS, Ryu MY, et al. Heterologous expression molecular and cellular characterization of CaPUB1 encoding a hot pepper U-Box E3 ubiquitin ligase homolog[J]. Plant Physiology, 2006, 142(4):1664-1682.
doi: 10.1104/pp.106.087965 URL |
| [57] |
Bhattacharyya S, Yu H, Mim C. Regulated protein turnover:snapshots of the proteasome in action[J]. Nature Reviews Molecular Cell Biology, 2014, 15:122-133.
doi: 10.1038/nrm3741 URL |
| [58] | Min HJ, Jung YJ, Kang BG, et al. CaPUB1, a hot pepper U-box E3 Ubiquitin ligase, confers enhanced cold stress tolerance and decreased drought stress tolerance in transgenic rice(Oryza sativa L.)[J]. Molecules & Cells, 2016, 39:250-257. |
| [59] |
Yan J, Wang J, Li Q, et al. AtCHIP, a U-box-containing E3 ubiquitin ligase, plays a critical role in temperature stress tolerance in Arabidopsis[J]. Plant Physiology, 2003, 132(2):861-869.
doi: 10.1104/pp.103.020800 URL |
| [60] |
Luo J, Shen G, Yan J, et al. AtCHIP functions as an E3 ubiquitin ligase of protein phosphatase 2A subunits and alters plant response to abscisic acid treatment[J]. The Plant Journal, 2006, 46(4):649-657.
doi: 10.1111/tpj.2006.46.issue-4 URL |
| [61] | Kang M, Lee S, Abdelmageed H, et al. Arabidopsis stress associated protein 9 mediates biotic and abiotic stress responsive ABA signaling via the proteasome pathway[J]. Plant, Cell & Environment, 2017, 40(5):702-716. |
| [62] | 乌凤章, 王贺新. 笃斯越橘响应低温和短光周期的蛋白质组分析[J]. 园艺学报, 2019, 46(2):265-279. |
| Wu FZ, Wang HX. Proteome analysis of response of Vaccinium uligiuosum L. seedlings to low-temperature and short photoperiod[J]. Acta Horticulturae Sinica, 2019, 46(2):265-279. | |
| [63] |
Jiao L, Zhang YL, Wu J, et al. A novel U-boxprotein gene from“Zuoshanyi”grapevine(Vitis amurensis Rupr. cv.)involved in cold responsive gene expression in Arabidopsis thanliana[J]. Plant Molecular Biology Reporter, 2015, 33(3):557-568.
doi: 10.1007/s11105-014-0783-4 URL |
| [1] | JIANG Lu-yuan, FENG Mei-jing, DU Yu-qing, DI Bao, CHEN Duan-fen, QIU De-you, YANG Yan-fang. Semi-lethal Low Temperature and Taxane Content of Taxus Under Low Temperature Stress [J]. Biotechnology Bulletin, 2023, 39(3): 232-242. |
| [2] | CUI Jun-mei, WEI Jia-ping, DONG Xiao-yun, WANG Ying, ZHENG Guo-qiang, LIU Zi-gang. PIP/PIPL: A Kind of Endogenous Plant Peptide Regulating Plant Stress Response and Development [J]. Biotechnology Bulletin, 2023, 39(3): 35-42. |
| [3] | CHEN Guang-xia, LI Xiu-jie, JIANG Xi-long, SHAN Lei, ZHANG Zhi-chang, LI Bo. Research Progress in Plant Small Signaling Peptides Involved in Abiotic Stress Response [J]. Biotechnology Bulletin, 2023, 39(11): 61-73. |
| [4] | MAO Ke-xin, WANG Hai-rong, AN Miao, LIU Teng-fei, WANG Shi-jin, LI Jian, LI Guo-tian. Identification of GRAS Gene Family and Expression Analysis Under Low Temperature Stress in Actinidia chinensis [J]. Biotechnology Bulletin, 2023, 39(11): 297-307. |
| [5] | YOU Chui-huai, XIE Jin-jin, ZHANG Ting, CUI Tian-zhen, SUN Xin-lu, ZANG Shou-jian, WU Yi-ning, SUN Meng-yao, QUE You-xiong, SU Ya-chun. Identification of the Lipoxygenase Gene GeLOX1 and Expression Analysis Under Low Temperature Stress in Gelsmium elegans [J]. Biotechnology Bulletin, 2023, 39(11): 318-327. |
| [6] | JIN Jiao-jiao, LIU Zi-gang, MI Wen-bo, XU Ming-xia, ZOU Ya, XU Chun-mei, ZHAO Cai-xia. Identification of Low Temperature Stress-responsive Genes Regulating Photosynthetic Characteristics in the Leaves of Brassica napus by RNA-Seq [J]. Biotechnology Bulletin, 2022, 38(4): 126-142. |
| [7] | LEI Chun-xia, LI Can-hui, CHEN Yong-kun, GONG Ming. Physiological and Biochemical Basis and Molecular Mechanism of Solanum tuberosum Tuberization [J]. Biotechnology Bulletin, 2022, 38(4): 44-57. |
| [8] | CUI Jie-bing, ZHANG Meng, ZHANG Ying-ting, XU Jin. Effects of Low Temperature Stress on Different Clones of Cryptomeria fortunei and Evaluation of Their Cold Resistance [J]. Biotechnology Bulletin, 2022, 38(3): 31-40. |
| [9] | LI Wen-jiao, ZHANG Zhong-feng, LIU Qing, SUN Jie, YANG Li, WANG Xing-jun, ZHAO Shu-zhen. Role of BRs in Plant Response to Abiotic Stress [J]. Biotechnology Bulletin, 2022, 38(1): 228-235. |
| [10] | WANG Lu-lu, GENG Xing-min, XU Shi-da. Ethylene Receptor in Fruit Ripening and Flower Senescence [J]. Biotechnology Bulletin, 2021, 37(3): 144-152. |
| [11] | LIU Jing, LI Ya-chao, ZHOU Meng-yan, WU Peng-fei, MA Xiang-qing, LI Ming. Advances in the Studies of Plant Protein Post-translational Modification [J]. Biotechnology Bulletin, 2021, 37(1): 67-76. |
| [12] | HU Xiao-qian, ZHANG Ying-yi, LI Xin, YAN Hai-fang. Research Progress of Remorin Protein in Plants [J]. Biotechnology Bulletin, 2020, 36(8): 136-143. |
| [13] | YANG Rui-jia, ZHANG Zhong-bao, WU Zhong-yi. Progress of the Structural and Functional Analysis of Plant Transcription Factor TIFY Protein Family [J]. Biotechnology Bulletin, 2020, 36(12): 121-128. |
| [14] | YU Ming-xiang, SONG Shui-shan. Biological Functions of Protein Fatty Acylation in Plant Cells [J]. Biotechnology Bulletin, 2019, 35(8): 170-177. |
| [15] | WANG Jia-yue, LIU Xiang-nan, PENG Kang-li, ZHAO Bo. Construction and Identification of Lentiviral Vector for RNA Interference of USE1 Gene [J]. Biotechnology Bulletin, 2019, 35(3): 117-122. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
