








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (9): 180-190.doi: 10.13560/j.cnki.biotech.bull.1985.2020-1419
Previous Articles Next Articles
CAO Ru-fei1( ), LI Ze-xuan2, XU Huan2, ZHANG Sha2, ZHANG Min-min2, DAI Feng2, DUAN Xiao-lei2,3(
), LI Ze-xuan2, XU Huan2, ZHANG Sha2, ZHANG Min-min2, DAI Feng2, DUAN Xiao-lei2,3( )
)
Received:2020-11-19
Online:2021-09-26
Published:2021-10-25
Contact:
DUAN Xiao-lei
E-mail:caofeier1221@126.com;duanxiaolei0@163.com
CAO Ru-fei, LI Ze-xuan, XU Huan, ZHANG Sha, ZHANG Min-min, DAI Feng, DUAN Xiao-lei. Expression,Purification,and Crystallization of Pif1 Helicase from Bacteroides fragilis[J]. Biotechnology Bulletin, 2021, 37(9): 180-190.

Fig. 1 Construction of expression vector pET15b-SUMO-B.f Pif1 A:The schematic map of pET15b-SUMO-B.f Pif1. B:Identification of the recombinant vector by PCR and restriction enzyme digestion. Lane 1,the product of colony PCR;lane 2,the digestion of control plasmid;lane 3,the double digestion of pET15b-SUMO-B.f Pif1 by Nde I and EcoR I;M:DNA DS5 000. Target bands indicated by red arrows

Fig. 2 Expression of B.f Pif1 protein induced with different IPTG concentrations and different induction temperatures A:SDS-PAGE of B.f Pif1 expression induced with different IPTG concentrations. 1-4 lanes were protein supernatants after induction with 0.1,0.3,0.5,and 0.8 mmol/L IPTG(lane 4 corresponds to the induced protein obscured by loading buffer change). B:SDS-PAGE of B.f pif1 expression induced with different induction temperatures, 1-3 lanes correspond to protein supernatants were inducted at 37℃ for 4 h,28℃ for 8 h,and 18℃ for 16 h,respectively(red arrows indicate target proteins of interest)
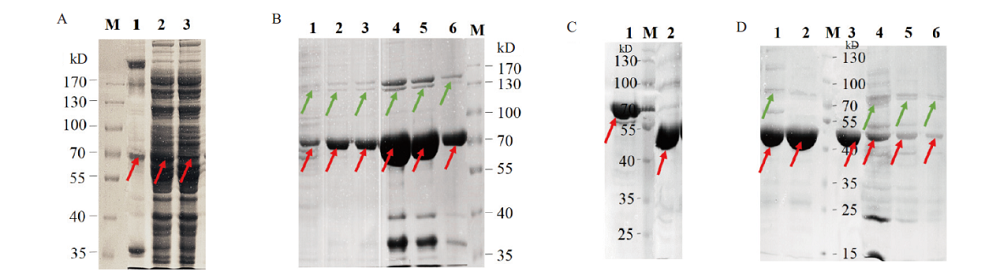
Fig. 3 Induced expression,Ni-NTA purification and SUMO digestion of B.f Pif1 protein A:SDS-PAGE analysis of the pellets,supernatants,and out flows of B.f Pif1 induced expression after bacterial ultrasonic crushing;B:SDS-PAGE of the first Ni-NTA purification with different concentrations of imidazole eluate buffer(1-2 lanes were wash tubes and 3-6 lanes were elute tubes,in which 4 and 5 lanes correspond to 300 mmol / L imidazole elution tubes;C:SDS-PAGE before and after SUMO digestion of the protein tags in B.f Pif1;D:SDS-PAGE of the second Ni-NTA purification with the supernatant and precipitate(red arrows indicated the target protein and green arrows indicated the hybrid protein)
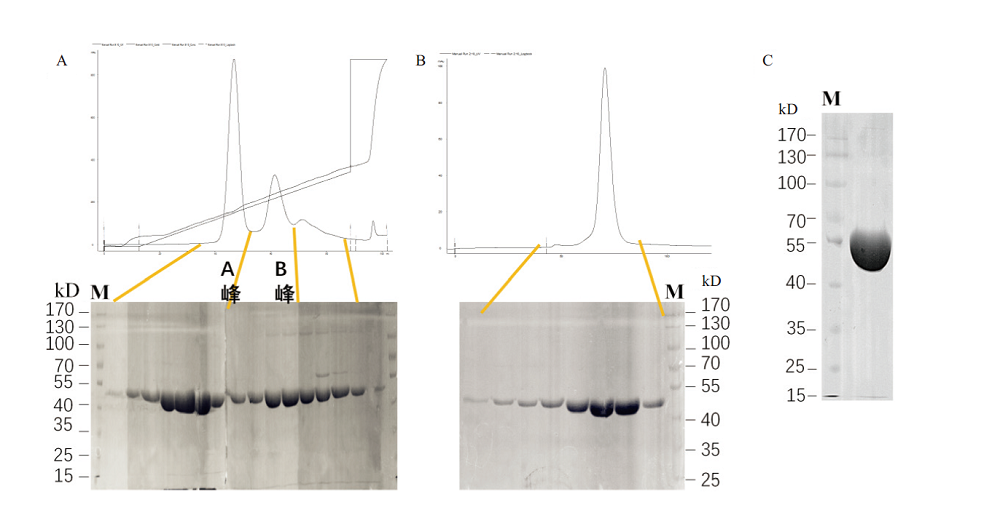
Fig. 4 Purifications of B.f Pif1 protein by DEAE ion-exchange chromatography and S200 gel filtration chromatography A:The absorption peak pattern and SDS-PAGE of the recombinant B.f Pif1 protein by DEAE ion-exchange chromatography. Peak A and B were eluted peaks at 2 different conductivities. B:The absorption peak pattern and SDS-PAGE of the recombinant B.f Pif1 protein by S200 gel filtration chromatography. C:SDS-PAGE of the final protein purified product with the concentration of 17 mg/mL by spotting 1μL of sample
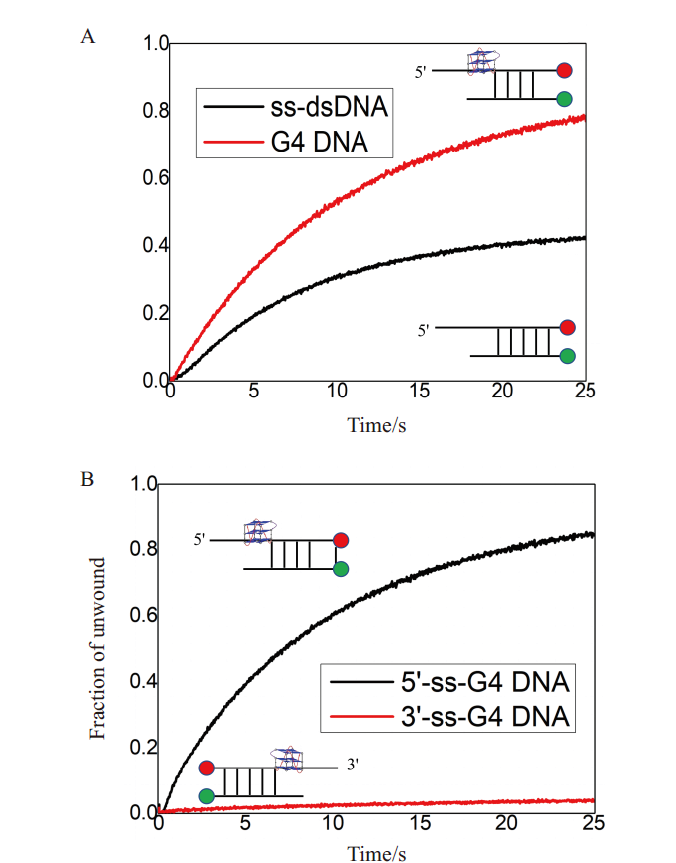
Fig. 5 Purified B.f Pif1 protein showing good biological activity A:Comparison of activity of B.f Pif1 protein unwinding G-quadruplex and ss-dsdNA substrate;B:B.f Pif1 helicase with good 5'-3' unwinding polarity

Fig. 6 Protein crystals of B.f Pif1 preliminarily screened with different crystallization kits A:Crystals were obtained in the 35th condition of the Salt RxTMⅠ kit;B:crystals were obtained in the 74th condition of the Cystal screen II kit;C:ultraviolet microscope photograph of the selected crystals in the 74th condition of the Cystal screen II kit with UY200 microscope,of which the green lines were plotting scales
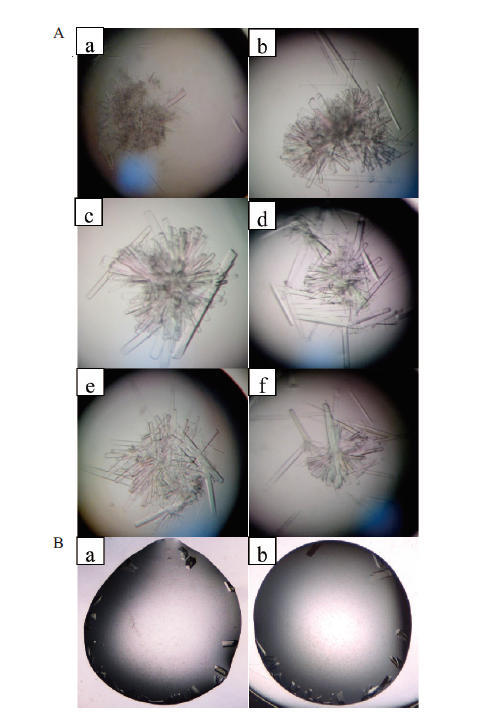
Fig. 7 Optimization of the culture conditions for B.f Pif1 protein crystals A:The gradual changes of B.f Pif1 twin-crystals were observed by microscope with hanging drop method;a-c subgraphs:gradient dilutions of B.f Pif1 protein 17 mg/mL(the original concentration),d-f subgraphs:the change of precipitants with different PEG and spermidine;B:the gradual formation of B.f Pif 1 single crystal were observed by microscope with the sitting drop method

Fig. 8 Growths of B.f Pif1 protein crystals A series of microscopic photographs of B.f Pif1 protein crystal growing status were observed in the same droplet with sitting drop method;the upper left corner was the initial observation time(unit:day),and the photos from top left to bottom right showed the state of the crystals on 0,3,5,8,13 and 19 d respectively
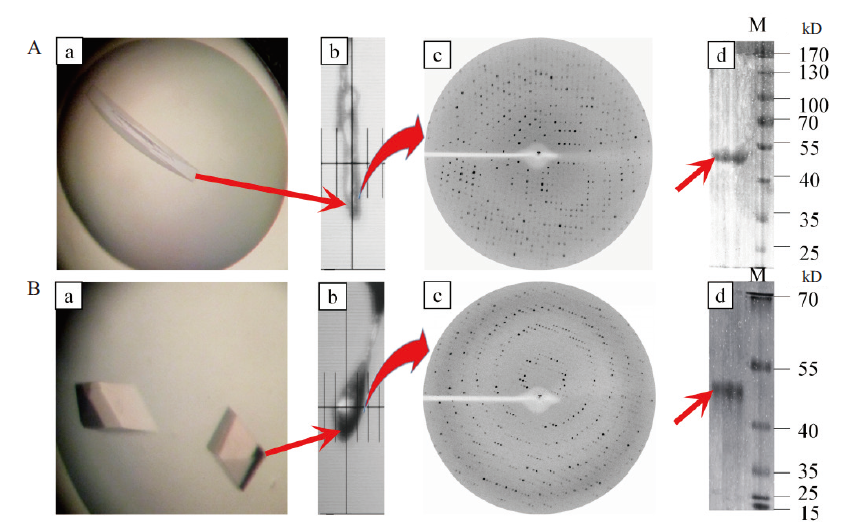
Fig. 9 X-ray diffraction patterns of B.f pif1 protein single crystal under different conditions and its corresponding SDS-PAGE A:X-ray diffraction and SDS-PAGE of the single protein crystal grown under the conditions of 100 mmol/L NH4Acetate,16% PEG4000 and pH 6.5;B:X-raydiffraction and SDS-PAGE of single crystals grown under the conditions of 0.1 mol/L mol/L Bis-Tris acetic acid(pH 8.3),0.05 mol/L sodium bicarbonate,5% glycerol and 0.015 mol / L spermidine
| [1] |
Sun F, Zhang Q, Zhao J, et al. A potential species of next-generation probiotics? The dark and light sides of Bacteroides fragilis in health[J]. Food Research International, 2019, 126(1):108590.
doi: 10.1016/j.foodres.2019.108590 URL |
| [2] |
Valguarnera E, Wardenburg JB. Good gone bad:One toxin away from disease for bacteroides fragilis[J]. Journal of Molecular Biology, 2020, 432(4):765-785.
doi: S0022-2836(19)30700-4 pmid: 31857085 |
| [3] |
Paul L, Patrick S, Nord CE, et al. The role of Bacteroides fragilis RecQ DNA helicases in cell survival after metronidazole exposure[J]. FEMS Microbiology Letter, 2011, 319(2):125-132.
doi: 10.1111/fml.2011.319.issue-2 URL |
| [4] |
Yaligara V, Fasahath H, Elizabeth LT, et al. Identification of genes required for the survival of Bfragilis using massive parallel sequencing of a saturated transposon mutant library[J]. BMC Genomics, 2014, 15(1):429-439.
doi: 10.1186/1471-2164-15-429 URL |
| [5] |
Tariq MA, Newberry F, Haagmans R, et al. Genome characterization of a novel wastewater bacteroides fragilis bacteriophage(vB_BfrS_23)and its host GB124[J]. Front Microbiol, 2020, 11(1):583378-583396.
doi: 10.3389/fmicb.2020.583378 URL |
| [6] |
Vacante M, Ciuni R, Basile F, et al. Gut microbiota and colorectal cancer development:A closer look to the adenoma-carcinoma sequence[J]. Biomedicines, 2020, 8(11):489-507.
doi: 10.3390/biomedicines8110489 URL |
| [7] |
Paeschke K, Bochman ML, Garcia PD, et al. Pif1 family helicases suppress genome instability at G-quadruplex motifs[J]. Nature, 2013, 497(7450):458-462.
doi: 10.1038/nature12149 URL |
| [8] |
Muellner J, Schmidt KH. Yeast genome maintenance by the multifunctional PIF1 DNA helicase family[J]. Genes, 2020, 11(2):224-245.
doi: 10.3390/genes11020224 URL |
| [9] |
Kuwahara T, Yamashita A, Hirakawa H, et al. Genomic analysis of Bacteroides fragilis reveals extensive DNA inversions regulating cell surface adaptation[J]. Proceedings of the National Academy of Science of the USA, 2004, 101(41):14919-14924.
doi: 10.1073/pnas.0404172101 URL |
| [10] |
Bannwarth S, Berg-Alonso L, Augé G, et al. Inactivation of Pif1 helicase causes a mitochondrial myopathy in mice[J]. Mitochondrion, 2016, 30(9):126-137.
doi: 10.1016/j.mito.2016.02.005 URL |
| [11] | Duan XL, Liu NN, Yang YT, et al. G-quadruplexes significantly stimulate Pif1 helicase-catalyzed duplex DNA unwinding[J]. Journal of Biological Chemistry, 2015, 292(12):7722-7735. |
| [12] |
Wei XB, Zhang B, Bazeille N, et al. A 3'-5' exonuclease activity embedded in the helicase core domain of Candida albicans Pif1 helicase[J]. Scientific Reports, 2017, 7(2):42865.
doi: 10.1038/srep42865 URL |
| [13] |
Chen WF, Dai YX, Duan XL, et al. Crystal structures of the BsPif1 helicase reveal that a major movement of the 2B SH3 domain is required for DNA unwinding[J]. Nucleic Acids Research, 2016, 44(6):2949-2961.
doi: 10.1093/nar/gkw033 URL |
| [14] |
Lu KY, Chen WF, Rety S, et al. Insights into the structural and mechanistic basis of multifunctional Scerevisiae Pif1p helicase[J]. Nucleic Acids Research, 2018, 46(3):1486-1500.
doi: 10.1093/nar/gkx1217 URL |
| [15] |
Saba DT, Vladimir L, Alfred AA, et al. Structural and functional analysis of the nucleotide and DNA binding activities of the human PIF1 helicase[J]. Nucleic Acids Research, 2019, 47(6):3208-3222.
doi: 10.1093/nar/gkz028 URL |
| [16] | 段晓雷, 刘娜女, 张涛, 等. 嗜热细菌解旋酶的异源表达与解旋反应特性[J]. 微生物学报, 2019, 59(3):566-577. |
| Duan XL, Liu NN, Zhang T, et al. Heterologous expression and characterization of unwinding reaction of Pif1 helicase from Anaerobaculum hydrogeniforman[J]. Acta Microbiologica Sinica, 2019, 59(3):566-577. | |
| [17] | 段晓雷, 许欢, 姚淼, 等. 嗜热厌氧杆菌解旋酶核心结构域蛋白与底物结合的反应特性[J]. 贵州医科大学学报, 2018, 43(11):35-44. |
| Duan XL, Xu H, Yao M, et al. Characteristic analysis of binding reactions of anaerobaculum hydrogeniforman Pif1 helicase’s core domain[J]. Journal of Guizhou Medical University, 2018, 43(11):35-44. | |
| [18] |
Chung L, Thiele OE, Geis AL, et al. Bacteroides fragilis toxin coordinates a pro-carcinogenic inflammatory cascade via targeting of colonic epithelial cells[J]. Cell Host Microbe, 2018, 23(2):203-214.
doi: S1931-3128(18)30042-8 pmid: 29398651 |
| [19] |
Mina Y, Hossein BB, Behrooz N, et al. To resist and persist:Important factors in the pathogenesis of Bacteroides fragilis[J]. Microbial Pathogenesis, 2020, 149(1):104506-104515.
doi: 10.1016/j.micpath.2020.104506 URL |
| [20] |
Gavira JA. Current trends in protein crystallization[J]. Archives of Biochemistry and Biophysics, 2016, 602(1):3-11.
doi: 10.1016/j.abb.2015.12.010 URL |
| [21] |
Marblestone JG, Edavettal SC, Lim Y, et al. Comparison of SUMO fusion technology with traditional gene fusion systems:enhanced expression and solubility with SUMO[J]. Protein Science, 2006, 15(1):182-189.
pmid: 16322573 |
| [22] |
Wang H, Xiao Y, Fu L, et al. High-level expression and purification of soluble recombinant FGF21 protein by SUMO fusion in Escherichia coli[J]. BMC Biotechnology, 2010, 10(1):14-17.
doi: 10.1186/1472-6750-10-14 URL |
| [23] |
Liu NN, Duan XL, Ai X, et al. The Bacteroides sp3_1_23 Pif1 protein is a multifunctional helicase[J]. Nucleic Acids Research, 2015, 43(18):8942-8954.
doi: 10.1093/nar/gkv916 URL |
| [24] |
Noirclerc-Savoye M, Flayhan A, Pereira C, et al. Tail proteins of phage T5:investigation of the effect of the His6-tag position, from expression to crystallisation[J]. Protein Expression and Purification, 2015, 109(1):70-78.
doi: 10.1016/j.pep.2015.02.003 URL |
| [25] | 黄嘉欣, 李海涛. 人组蛋白α-氨基乙酰基转移酶Nat11表达纯化、晶体生长及底物结合研究[J]. 生物技术通报, 2014, 30(11):193-200. |
| Huang JX, Li HT. Expression, crystallization and substrate binding studies of human histone N-terminal acetyltransferase Nat11[J]. Biotechnology Bulletin, 2014, 30(11):193-200. | |
| [26] | Sparks MA, Singh SP, Burgers PM, et al. Complementary roles of Pif1 helicase and single stranded DNA binding proteins in stimulating DNA replication through G-quadruplexes[J]. Nucleic Acids Research, 2019, 47(16):8595-8605. |
| [27] | 林波, 刘坤, 朱明月, 等. α-芋螺毒素GIC与Ac-AChBP共结晶条件的筛选与优化[J]. 生物技术通报, 2017, 33(2):192-198. |
| Lin B, Liu K, Zhu MY, et al. Screening and optimization of Co-crystallization condition of α-conotoxin GIC complex with Ac-AchBP[J]. Biotechnology Bulletin, 2017, 33(2):192-198. | |
| [28] |
Erturk-Hasdemir D, Kasper DL. Finding a needle in a haystack:Bacteroides fragilis polysaccharide A as the archetypical symbiosis factor[J]. Annals of The New York Academy of Sciences, 2018, 1417(1):116-129.
doi: 10.1111/nyas.13660 pmid: 29528123 |
| No related articles found! |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||