








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (10): 57-62.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0036
Previous Articles Next Articles
CUI Xin-gang1( ), SUN Ya-xin1, CUI Xiao-jing2, DENG Yan-wen3, SUN En-hao3, WANG Jun-fang4, CUI Hong-jing2,4(
), SUN Ya-xin1, CUI Xiao-jing2, DENG Yan-wen3, SUN En-hao3, WANG Jun-fang4, CUI Hong-jing2,4( )
)
Received:2021-01-10
Online:2021-10-26
Published:2021-11-12
Contact:
CUI Hong-jing
E-mail:dic456@163.com;cuihongjing@gdmu.edu.cn
CUI Xin-gang, SUN Ya-xin, CUI Xiao-jing, DENG Yan-wen, SUN En-hao, WANG Jun-fang, CUI Hong-jing. Roles of Gene TAP42 in the Cell Wall Stress Response of Saccharomyces cerevisiae[J]. Biotechnology Bulletin, 2021, 37(10): 57-62.
| 引物名称Primer name | 引物序列Primer Sequence(5'-3') | 产物大小 Amplicon size/bp | 用途Purpose |
|---|---|---|---|
| TAP42-F | AAGAAGAGGAAGAAAGTAATAACGAGGAGAGATCATAAACCTGTGCGGTATTTCACACCG | 1 232 | 扩增基因 破坏元件 |
| TAP42-R | TCTAATAGATACTTATGTGTACGTATATAGGTATTTTTTTAGATTGTACTGAGAGTGCAC | ||
| V-TAP42-F | TGGTGGTGTTGTAGCGGTAAATG | 1 678 | 扩增基因 破坏片段 |
| V-TAP42-R | TCTCGGAAAGAAAAGAGCCTGTG | ||
| TAP42-int-F | CCCTCAGCTTAGCGAATTGTAC | 164 | 扩增目的 基因内部片段 |
| TAP42-int-R | CGTGGTCCTCGTCGTTGTTT | ||
| TAP42-int-F | CCCTCAGCTTAGCGAATTGTAC | 712 | 扩增破坏 元件部分片段 |
| URA-int-F | GGAACCTAGAGGCCTTTTGATGT | ||
| SWI4-int-F | AGGTGGGTATGGTAGGT | 254 | 定量引物 |
| SWI4-int-R | TTGTAGCCGATGTTGAT | ||
| SWI6-int-F | GCAAGCCTAACGATGAT | 214 | 定量引物 |
| SWI6-int-R | CGTTGTAAGAATGCCTCTA | ||
| SSD1-int-F | CTGTTGATAGGCGTGCGAGA | 143 | 定量引物 |
| SSD1-int-R | AACAACCGACAGCGTGGC | ||
| MPT5-int-F | ACGCATCTCAGGCCACCTG | 194 | 定量引物 |
| MPT5-int-R | GATGCTGAGTCGGCAGATGG |
Table 1 Nucleotide sequence of primers used in this study
| 引物名称Primer name | 引物序列Primer Sequence(5'-3') | 产物大小 Amplicon size/bp | 用途Purpose |
|---|---|---|---|
| TAP42-F | AAGAAGAGGAAGAAAGTAATAACGAGGAGAGATCATAAACCTGTGCGGTATTTCACACCG | 1 232 | 扩增基因 破坏元件 |
| TAP42-R | TCTAATAGATACTTATGTGTACGTATATAGGTATTTTTTTAGATTGTACTGAGAGTGCAC | ||
| V-TAP42-F | TGGTGGTGTTGTAGCGGTAAATG | 1 678 | 扩增基因 破坏片段 |
| V-TAP42-R | TCTCGGAAAGAAAAGAGCCTGTG | ||
| TAP42-int-F | CCCTCAGCTTAGCGAATTGTAC | 164 | 扩增目的 基因内部片段 |
| TAP42-int-R | CGTGGTCCTCGTCGTTGTTT | ||
| TAP42-int-F | CCCTCAGCTTAGCGAATTGTAC | 712 | 扩增破坏 元件部分片段 |
| URA-int-F | GGAACCTAGAGGCCTTTTGATGT | ||
| SWI4-int-F | AGGTGGGTATGGTAGGT | 254 | 定量引物 |
| SWI4-int-R | TTGTAGCCGATGTTGAT | ||
| SWI6-int-F | GCAAGCCTAACGATGAT | 214 | 定量引物 |
| SWI6-int-R | CGTTGTAAGAATGCCTCTA | ||
| SSD1-int-F | CTGTTGATAGGCGTGCGAGA | 143 | 定量引物 |
| SSD1-int-R | AACAACCGACAGCGTGGC | ||
| MPT5-int-F | ACGCATCTCAGGCCACCTG | 194 | 定量引物 |
| MPT5-int-R | GATGCTGAGTCGGCAGATGG |
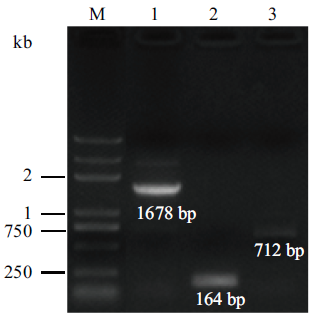
Fig. 1 Agarose gel electrophoresis of PCR products on verifying the TAP42 deletion strain M represents DNA marker;the bands of lane 1 and 3 are the full or part length PCR products of disruption module as template of tap42Δ genomic DNA;and the band of lane 2 is PCR products of an internal fragment from the TAP42 gene.

Fig. 2 Cell proliferation ability of the yeast strains The colony-forming ability (A) and cell proliferation activity (B) of BY4743 and tap42Δ strains. BY4743 is the control wild strain
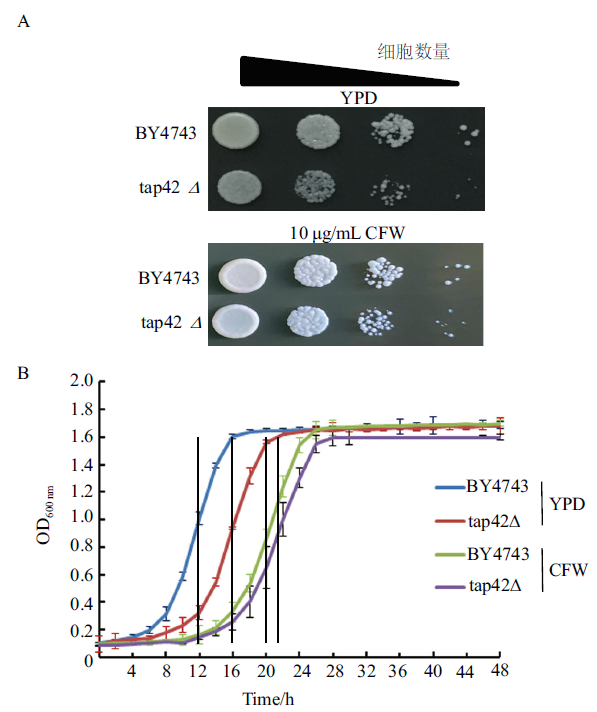
Fig.3 Resistance to the cell wall stress response of the yeast strains The colony-forming ability (A) and cell proliferation activity (B) of BY4743 and tap42Δ strains without or with 10 μg/mL calcofluor white stain (CFW) treatment. BY4743 is the control wild strain

Fig.4 Expression levels of SWI4, SWI6, MPT5 and SSD genes in yeast strains The relative mRNA expression levels of SWI4, SWI6, SSD1 and MPT5 genes for BY4743 and tap42Δ strains. Data are in the form of mean±standard error. *: P < 0.05, **: P<0.01 vs BY4743; CFW is calcofluor white stain as an inducer of cell wall stress response. BY4743 is the control wild strain
| [1] |
Levin DE. Regulation of cell wall biogenesis in Saccharomyces cerevisiae:the cell wall integrity signaling pathway[J]. Genetics. 2011, 189(4):1145-1175.
doi: 10.1534/genetics.111.128264 URL |
| [2] |
Kaeberlein M, Guarente L. Saccharomyces cerevisiae MPT5 and SSD1 function in parallel pathways to promote cell wall integrity[J]. Genetics, 2002, 160(1):83-95.
pmid: 11805047 |
| [3] |
Kaeberlein M, Andalis AA, Liszt GB, et al. Saccharomyces cerevisiae SSD1-V confers longevity by a Sir2p-independent mechanism[J]. Genetics, 2004, 166(4):1661-1672.
pmid: 15126388 |
| [4] |
Di Como CJ, Arndt KT. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases[J]. Genes and Development, 1996, 10(15):1904-1916.
pmid: 8756348 |
| [5] |
Reinke A, Anderson S, McCaffery JM, et al. TOR complex 1 includes a novel component, Tco89p(YPL180w), and cooperates with Ssd1p to maintain cellular integrity in Saccharomyces cerevisiae[J]. Journal of Biological Chemistry, 2004, 279(15):14752-14762.
pmid: 14736892 |
| [6] |
Cui HJ, Cui XG, Zhao W, et al. PMT1 deficiency extends the shortened replicative lifespan of TED1-deficient yeast in a Hac1p-dependent manner[J]. FEMS Microbiology Letters, 2018, 365(21). DOI: 10.1093/femsle/fny234.
doi: 10.1093/femsle/fny234 |
| [7] |
Zhao W, Zheng HZ, Zhou T, et al. CTT1 overexpression increases the replicative lifespan of MMS-sensitive Saccharomyces cerevisiae deficient in KSP1[J]. Mechanisms of Ageing and Development, 2017, 164:27-36.
doi: S0047-6374(16)30224-X pmid: 28347693 |
| [8] |
Kumar A, John L, Alam MM, et al. Homocysteine- and cysteine-mediated growth defect is not associated with induction of oxidative stress response genes in yeast[J]. Biochemical Journal, 2006, 396(1):61-69.
pmid: 16433631 |
| [9] |
Hung CW, Martinez-Marquez JY, Javed FT, et al. A simple and inexpensive quantitative technique for determining chemical sensitivity in Saccharomyces cerevisiae[J]. Scientific Reports, 2018, 8(1):11919.
doi: 10.1038/s41598-018-30305-z URL |
| [10] |
Zhao W, Fang BX, Niu YJ, et al. Nar1 deficiency results in shortened lifespan and sensitivity to paraquat that is rescued by increased expression of mitochondrial superoxide dismutase[J]. Mechanisms of Ageing and Development, 2014, 138:53-58.
doi: 10.1016/j.mad.2014.01.007 pmid: 24486555 |
| [11] | Cui HJ, Liu XG, McCormick M, et al. PMT1 deficiency enhances basal UPR activity and extends replicative lifespan of Saccharomyces cerevisiae[J]. Age(Dordr), 2015, 37(3):9788. |
| [12] |
Cygnar KD, Gao X, Pan D, et al. The phosphatase subunit tap42 functions independently of target of rapamycin to regulate cell division and survival in Drosophila[J]. Genetics, 2005, 170(2):733-740.
doi: 10.1534/genetics.104.039909 URL |
| [13] |
Duvel K, Santhanam A, Garrett S, et al. Multiple roles of Tap42 in mediating rapamycin-induced transcriptional changes in yeast[J]. Molecular Cell, 2003, 11(6):1467-1478.
doi: 10.1016/S1097-2765(03)00228-4 URL |
| [14] |
Yorimitsu T, He C, Wang K, et al. Tap4-associated protein phosphatase type 2A negatively regulates induction of autophagy[J]. Autophagy, 2009, 5(5):616-624.
pmid: 19223769 |
| [15] |
Liu Y, Okamoto K. The TORC1 signaling pathway regulates respiration-induced mitophagy in yeast[J]. Biochemical and Biophysical Research Communications, 2018, 502(1):76-83.
doi: 10.1016/j.bbrc.2018.05.123 URL |
| [16] |
Wang N, Leung HT, Mazalouskas MD, et al. Essential roles of the Tap42-regulated protein phosphatase 2A(PP2A)family in wing Imaginal disc development of Drosophila melanogaster[J]. PLoS One, 2012: 7(6):e38569.
doi: 10.1371/journal.pone.0038569 URL |
| [17] |
Ling NXY, Kaczmarek A, Hoque A, et al. mTORC1 directly inhibits AMPK to promote cell proliferation under nutrient stress[J]. Nature Metabolism, 2020, 2(1):41-49.
doi: 10.1038/s42255-019-0157-1 URL |
| [18] | Sanz AB, Garcia R, Rodriguez-Pena JM, et al. Slt2 MAPK association with chromatin is required for transcriptional activation of Rlm1 dependent genes upon cell wall stress[J]. Biochimica et Biophysica Acta(BBA)-Gene Regulatory Mechanisms, 2018, 1861(11):1029-1039. |
| [1] | HUANG Xiao-long, SUN Gui-lian, MA Dan-dan, YAN Hui-qing. Construction of Yeast One-hybrid Library and Screening of Factors Regulating LAZY1 Expression in Rice [J]. Biotechnology Bulletin, 2023, 39(9): 126-135. |
| [2] | WEN Xiao-lei, LI Jian-yuan, LI Na, ZHANG Na, YANG Wen-xiang. Construction and Utilization of Yeast Two-hybrid cDNA Library of Wheat Interacted by Puccinia triticina [J]. Biotechnology Bulletin, 2023, 39(9): 136-146. |
| [3] | HAN Hao-zhang, ZHANG Li-hua, LI Su-hua, ZHAO Rong, WANG Fang, WANG Xiao-li. Construction of cDNA Library of Cinnamomun bodinieri Induced by Saline-alkali Stress and Screening of CbP5CS Upstream Regulators [J]. Biotechnology Bulletin, 2023, 39(9): 236-245. |
| [4] | XU Fa-di, XU Kang, SUN Dong-ming, LI Meng-lei, ZHAO Jian-zhi, BAO Xiao-ming. Research Progress in Second-generation Fuel Ethanol Technology Based on Poplar(Populus sp.) [J]. Biotechnology Bulletin, 2023, 39(9): 27-39. |
| [5] | JIANG Run-hai, JIANG Ran-ran, ZHU Cheng-qiang, HOU Xiu-li. Research Progress in Mechanisms of Microbial-enhanced Phytoremediation for Lead-contaminated Soil [J]. Biotechnology Bulletin, 2023, 39(8): 114-125. |
| [6] | ZHAO Si-jia, WANG Xiao-lu, SUN Ji-lu, TIAN Jian, ZHANG Jie. Modification of Pichia pastoris for Erythritol Production by Metabolic Engineering [J]. Biotechnology Bulletin, 2023, 39(8): 137-147. |
| [7] | LYU Qiu-yu, SUN Pei-yuan, RAN Bin, WANG Jia-rui, CHEN Qing-fu, LI Hong-you. Cloning, Subcellular Localization and Expression Analysis of the Transcription Factor Gene FtbHLH3 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 194-203. |
| [8] | XU Jing, ZHU Hong-lin, LIN Yan-hui, TANG Li-qiong, TANG Qing-jie, WANG Xiao-ning. Cloning of IbHQT1 Promoter and Identification of Upstream Regulatory Factors in Sweet Potato [J]. Biotechnology Bulletin, 2023, 39(8): 213-219. |
| [9] | LI Bo, LIU He-xia, CHEN Yu-ling, ZHOU Xing-wen, ZHU Yu-lin. Cloning, Subcellular Localization and Expression Analysis of CnbHLH79 Transcription Factor from Camellia nitidissima [J]. Biotechnology Bulletin, 2023, 39(8): 241-250. |
| [10] | CHEN Xiao, YU Ming-lan, WU Long-kun, ZHENG Xiao-ming, PANG Hong-bo. Research Progress in lncRNA and Their Responses to Low Temperature Stress in Plant [J]. Biotechnology Bulletin, 2023, 39(7): 1-12. |
| [11] | HU Hai-lin, XU Li, LI Xiao-xu, WANG Chen-can, MEI Man, DING Wen-jing, ZHAO Yuan-yuan. Advances in the Regulation of Plant Growth, Development and Stress Physiology by Small Peptide Hormones [J]. Biotechnology Bulletin, 2023, 39(7): 13-25. |
| [12] | YOU Ling, JIAN Xiao-ping, FAN Fang-yong, YANG Zhi, WANG Tao. Ecological Monitoring of Pit Mud in Yibin Strong-fragrance Baijiu-producing Region [J]. Biotechnology Bulletin, 2023, 39(7): 254-265. |
| [13] | CHENG Ting, YUAN Shuai, ZHANG Xiao-yuan, LIN Liang-cai, LI Xin, ZHANG Cui-ying. Research Progress in the Regulation of Isobutanol Synthesis Pathway in Saccharomyces cerevisiae [J]. Biotechnology Bulletin, 2023, 39(7): 80-90. |
| [14] | YOU Zi-juan, CHEN Han-lin, DENG Fu-cai. Research Progress in the Extraction and Functional Activities of Bioactive Peptides from Fish Skin [J]. Biotechnology Bulletin, 2023, 39(7): 91-104. |
| [15] | CHEN Cai-ping, REN Hao, LONG Teng-fei, HE Bing, LU Zhao-xiang, SUN Jian. Research Advances in the Treatment of Inflammation Bowel Disease Using Escherichia coli Nissle 1917 [J]. Biotechnology Bulletin, 2023, 39(6): 109-118. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||