








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (6): 279-285.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0231
Previous Articles Next Articles
WANG Jing-wen1( ), YAN Fang1, LIU Lang1, ZHOU Xue-ping1,2, WANG Dao-wen3,4, ZHOU Huan-bin1,5(
), YAN Fang1, LIU Lang1, ZHOU Xue-ping1,2, WANG Dao-wen3,4, ZHOU Huan-bin1,5( )
)
Received:2020-03-01
Online:2021-06-26
Published:2021-07-08
Contact:
ZHOU Huan-bin
E-mail:wangjingwenHN@163.com;hbzhou@ippcaas.cn
WANG Jing-wen, YAN Fang, LIU Lang, ZHOU Xue-ping, WANG Dao-wen, ZHOU Huan-bin. Optimization of CRISPR/Cas12a System and Development of It-mediated Adenine Base Editor in Rice[J]. Biotechnology Bulletin, 2021, 37(6): 279-285.
| Name | Source | |
|---|---|---|
| Plasmid | pUC57:LbCas12a-crRNA | Bought from Sangon Biotech |
| pUC57:AsCas12a-crRNA | Bought from Sangon Biotech | |
| pUC57:LbCas12a | Bought from Sangon Biotech | |
| pUC57:AsCas12a | Bought from Sangon Biotech | |
| pENTR4 | Lab stock | |
| pHZ11 | This study | |
| pHZ4 | This study | |
| pUC19:rBE3 | Lab stock | |
| pUC19:LbCas12a | This study | |
| pUC19:AsCas12a | This study | |
| pUbi:rBE3 | Lab stock | |
| pUbi:LbCas12a | This study | |
| pUbi:AsCas12a | This study | |
| pUbi:rBE58 | This study | |
| Strain | JM109 | Lab stock |
| DB3.1 | Lab stock | |
| EHA105 | Lab stock |
Table 1 Plasmids and strains involved in this study
| Name | Source | |
|---|---|---|
| Plasmid | pUC57:LbCas12a-crRNA | Bought from Sangon Biotech |
| pUC57:AsCas12a-crRNA | Bought from Sangon Biotech | |
| pUC57:LbCas12a | Bought from Sangon Biotech | |
| pUC57:AsCas12a | Bought from Sangon Biotech | |
| pENTR4 | Lab stock | |
| pHZ11 | This study | |
| pHZ4 | This study | |
| pUC19:rBE3 | Lab stock | |
| pUC19:LbCas12a | This study | |
| pUC19:AsCas12a | This study | |
| pUbi:rBE3 | Lab stock | |
| pUbi:LbCas12a | This study | |
| pUbi:AsCas12a | This study | |
| pUbi:rBE58 | This study | |
| Strain | JM109 | Lab stock |
| DB3.1 | Lab stock | |
| EHA105 | Lab stock |

Fig.1 Schematics of gene editing constructs A:Schematics of Cas12a expression components. B:Schematics of crRNA vector. C:Schematics of the adenine base editor(pUbi:rBE58)
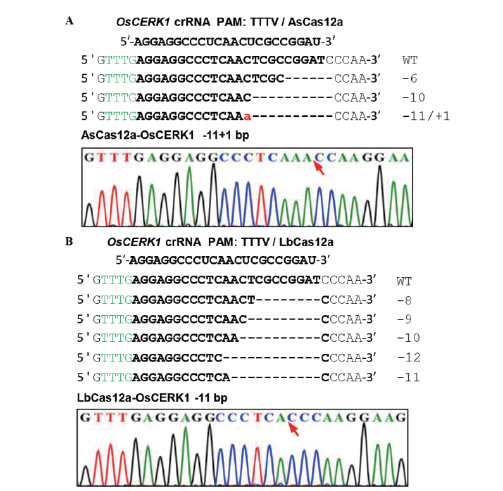
Fig.2 Detection of CRISPR/LbCas12a in rice protoplasts and the nuclease activity of CRISPR/AsCas12a A:PCR/RE detection of editing OsCERK1 gene in the rice protoplasts mediated by CRISPR/AsCas12a system. B:PCR/RE detection of editing OsCERK1 gene in the rice protoplasts mediated by CRISPR/LbCas12a system
| Gene name | Target site sequence |
|---|---|
| OsCERK1 | AGGAGGCCCTCAACTCGCCGGAT |
| OsCPK13 | ATGGGCAACGCATGCGGCGGTTC |
| OsCPK17 | ATAGGATCGTGCAGCGCGGGCAC |
| OsCPK15 | CACCACTAGATATAGAAGGCCAT |
| Bsrd1 | AATTTTAACTGATAAATATAAGT |
| OsJAZ4 | AGGACGAGATCATGGAGTCTGAC |
Table 2 Target sites used in this study
| Gene name | Target site sequence |
|---|---|
| OsCERK1 | AGGAGGCCCTCAACTCGCCGGAT |
| OsCPK13 | ATGGGCAACGCATGCGGCGGTTC |
| OsCPK17 | ATAGGATCGTGCAGCGCGGGCAC |
| OsCPK15 | CACCACTAGATATAGAAGGCCAT |
| Bsrd1 | AATTTTAACTGATAAATATAAGT |
| OsJAZ4 | AGGACGAGATCATGGAGTCTGAC |

Fig.4 Targeted adenine base editing of endogenous gene OsCPK15 in rice using the rBE58 system A:Various editing types of target site OsCPK15 generated by rBE58,target sequences,PAM sequences,targeted base T in the putative editing window,and detected A-to-G conversions are highlighted in bold,green,red,and blue,respectively;and the detected mutated nucleotide in the figure are indicated by the red underlines. B:Summary of target base editing efficiency in the activity window of base editing for the target sites ofOsCPK15 gene mediated by rBE58
| [1] | Makarova KS, Koonin EV. Annotation and classification of CRISPR-cas systems[J]. Methods Mol Biol, 2015, 1311:47-75. |
| [2] |
Koonin EV, Makarova KS, Zhang F. Diversity, classification and evolution of CRISPR-Cas systems[J]. Curr Opin Microbiol, 2017, 37:67-78.
doi: 10.1016/j.mib.2017.05.008 URL |
| [3] |
Makarova KS, Wolf YI, Iranzo J, et al. Evolutionary classification of CRISPR-Cas systems:a burst of class 2 and derived variants[J]. Nat Rev Microbiol, 2020, 18(2):67-83.
doi: 10.1038/s41579-019-0299-x URL |
| [4] |
Jinek M, Chylinski K, Fonfara I, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity[J]. Science, 2012, 337(6096):816-821.
doi: 10.1126/science.1225829 URL |
| [5] |
Zetsche B, Gootenberg JS, Abudayyeh OO, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system[J]. Cell, 2015, 163(3):759-771.
doi: 10.1016/j.cell.2015.09.038 pmid: 26422227 |
| [6] |
Strecker J, Jones S, Koopal B, et al. Engineering of CRISPR-Cas12b for human genome editing[J]. Nat Commun, 2019, 10(1):212.
doi: 10.1038/s41467-018-08224-4 URL |
| [7] |
Zetsche B, Heidenreich M, Mohanraju P, et al. Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array[J]. Nat Biotechnol, 2017, 35(1):31-34.
doi: 10.1038/nbt.3737 pmid: 27918548 |
| [8] |
Hur JK, Kim K, Been KW, et al. Targeted mutagenesis in mice by electroporation of Cpf1 ribonucleoproteins[J]. Nat Biotechnol, 2016, 34(8):807-808.
doi: 10.1038/nbt.3596 URL |
| [9] |
Endo A, Masafumi M, Kaya H, et al. Efficient targeted mutagenesis of rice and tobacco genomes using Cpf1 from Francisella novicida[J]. Sci Rep, 2016, 6:38169.
doi: 10.1038/srep38169 URL |
| [10] |
Tang X, Lowder LG, Zhang T, et al. A CRISPR-Cpf1 system for efficient genome editing and transcriptional repression in plants[J]. Nat Plants, 2017, 3:17018.
doi: 10.1038/nplants.2017.18 pmid: 28211909 |
| [11] | Wang M, Mao Y, Lu Y, et al. Multiplex gene editing in rice using the CRISPR-Cpf1 system[J]. Mol Plant, 2017, 7:1011-1013. |
| [12] |
Kim H, Kim ST, Ryu J, et al. CRISPR/Cpf1-mediated DNA-free plant genome editing[J]. Nat Commun, 2017, 8:14406.
doi: 10.1038/ncomms14406 URL |
| [13] |
Kim D, Lim K, Kim ST, et al. Genome-wide target specificities of CRISPR RNA-guided programmable deaminases[J]. Nat Biotechnol, 2017, 35(5):475-480.
doi: 10.1038/nbt.3852 URL |
| [14] | Van Vu T, Sivankalyani V, Kim EJ, et al. Homology-directed repair using next-generation CRISPR/Cpf1-geminiviral replicons in tomato[J]. bioRxiv, 2019: 521419. |
| [15] |
Malzahn AA, Tang X, Lee K, et al. Application of CRISPR-Cas12a temperature sensitivity for improved genome editing in rice, maize, and Arabidopsis[J]. BMC Biol, 2019, 17(1):9.
doi: 10.1186/s12915-019-0629-5 pmid: 30704461 |
| [16] |
Li J, Xu R, Qin R, et al. Genome editing mediated by SpCas9 variants with broad non-canonical PAM compatibility in plants[J]. Mol Plant, 2021, 14(2):352-360.
doi: 10.1016/j.molp.2020.12.017 URL |
| [17] |
Zhou H, Liu B, Weeks DP, et al. Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice[J]. Nucleic Acids Res, 2014, 42(17):10903-10914.
doi: 10.1093/nar/gku806 URL |
| [18] |
Ren B, Yan F, Kuang YJ, et al. A CRISPR/Cas9 toolkit for efficient targeted base editing to induce genetic variations in rice[J]. Sci China Life Sci, 2017, 60(5):516-519.
doi: 10.1007/s11427-016-0406-x URL |
| [19] |
Jiang W, Zhou H, Bi H, et al. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, Sorghum and rice[J]. Nucleic Acids Res, 2013, 41(20):e188.
doi: 10.1093/nar/gkt780 URL |
| [20] |
Hiei Y, Ohta S, Komari T, et al. Efficient transformation of rice(Oryza sativa L.)mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA[J]. The Plant Journal, 1994, 6(2):271-282.
doi: 10.1046/j.1365-313X.1994.6020271.x URL |
| [21] |
Moon SB, Lee JM, Kang JG, et al. Highly efficient genome editing by CRISPR-Cpf1 using CRISPR RNA with a uridinylate-rich 3'-overhang[J]. Nat Commun, 2018, 9(1):3651.
doi: 10.1038/s41467-018-06129-w pmid: 30194297 |
| [22] |
Li X, Wang Y, Liu Y, et al. Base editing with a Cpf1-cytidine deaminase fusion[J]. Nat Biotechnol, 2018, 36(4):324-327.
doi: 10.1038/nbt.4102 URL |
| [1] | HUANG Xiao-long, SUN Gui-lian, MA Dan-dan, YAN Hui-qing. Construction of Yeast One-hybrid Library and Screening of Factors Regulating LAZY1 Expression in Rice [J]. Biotechnology Bulletin, 2023, 39(9): 126-135. |
| [2] | LI Xue-qi, ZHANG Su-jie, YU Man, HUANG Jin-guang, ZHOU Huan-bin. Establishment of CRISPR/CasX-based Genome Editing Technology in Rice [J]. Biotechnology Bulletin, 2023, 39(9): 40-48. |
| [3] | LIU Jia-hui, LIU Ye, HUA Er-bing, WANG Meng. PAM Extension of Cytosine Base Editing Tool in Corynebacterium glutamicum [J]. Biotechnology Bulletin, 2023, 39(9): 49-57. |
| [4] | CHEN Xiao-ling, LIAO Dong-qing, HUANG Shang-fei, CHEN Ying, LU Zhi-long, CHEN Dong. Advances in CRISPR/Cas9 System Modifying Saccharomycescerevisiae [J]. Biotechnology Bulletin, 2023, 39(8): 148-158. |
| [5] | YANG Yu-mei, ZHANG Kun-xiao. Establishing a Stable Cell Line with Site-specific Integration of ERK Kinase Phase-separated Fluorescent Probe Using CRISPR/Cas9 Technology [J]. Biotechnology Bulletin, 2023, 39(8): 159-164. |
| [6] | SHI Wei-tao, YAO Chun-peng, WEI Wen-Kang, WANG Lei, FANG Yuan-jie, TONG Yu-jie, MA Xiao-jiao, JIANG Wen, ZHANG Xiao-ai, SHAO Wei. Establishment of MDH2 Knockout Cell Line Using CRISPR/Cas9 Technology and Study of Anti-deoxynivalenol Effect [J]. Biotechnology Bulletin, 2023, 39(7): 307-315. |
| [7] | ZHANG Zu-lin, LIU Fang-fang, ZHOU Qing-niao, ZHAO Rui-qiang, HE Shu-jia, LIN Wen-zhen. Construction and Identification of Huh7 Hepatoma Cell Line with ACE2 Gene Knockout Based on CRISPR/Cas9 Technology [J]. Biotechnology Bulletin, 2023, 39(6): 181-188. |
| [8] | LIU Xiao-yan, ZHU Zhen-liang, SHI Guang-yu, HUA Zi-yu, YANG Chen, ZHANG Yong, LIU Jun. Strategies to Optimize the Expression of Mammary Gland Bioreactor [J]. Biotechnology Bulletin, 2023, 39(5): 77-91. |
| [9] | LI Yi-jun, WU Chen-chen, LI Rui, WANG Zhe, HE Shan-wen, WEI Shan-jun, ZHANG Xiao-xia. Exploring Cultivation Approaches for New Endophytic Bacterial Resource in Oryza sativa [J]. Biotechnology Bulletin, 2023, 39(4): 201-211. |
| [10] | ZHOU Xiao-jie, YANG Si-qi, ZHANG Yi-wen, XU Jia-qi, YANG Sheng. CRISPR-associated Transposases and Their Applications in Bacterial Genome Editing [J]. Biotechnology Bulletin, 2023, 39(4): 49-58. |
| [11] | CHENG Jing-wen, CAO Lei, ZHANG Yan-min, YE Qian, CHEN Min, TAN Wen-song, ZHAO Liang. Establishment and Application of Multigene Engineering Transformation Strategy for CHO Cells [J]. Biotechnology Bulletin, 2023, 39(2): 283-291. |
| [12] | LU Zhen-wan, LI Xue-qi, HUANG Jin-guang, ZHOU Huan-bin. Creation of Glyphosate-tolerant Rice by Cytosine Base Editing [J]. Biotechnology Bulletin, 2023, 39(2): 63-69. |
| [13] | HUANG Wen-li, LI Xiang-xiang, ZHOU Wen-ting, LUO Sha, YAO Wei-jia, MA Jie, ZHANG Fen, SHEN Yu-sen, GU Hong-hui, WANG Jian-sheng, SUN Bo. Targeted Editing of BoZDS in Broccoli by CRISPR/Cas9 Technology [J]. Biotechnology Bulletin, 2023, 39(2): 80-87. |
| [14] | WANG Bing, ZHAO Hui-na, YU Jing, CHEN Jie, LUO Mei, LEI Bo. Regulation of Leaf Bud by REVOLUTA in Tobacco Based on CRISPR/Cas9 System [J]. Biotechnology Bulletin, 2023, 39(10): 197-208. |
| [15] | LI Shuang-xi, HUA Jin-lian. Research Progress in Anti-porcine Reproductive and Respiratory Syndrome Genetically Modified Pigs [J]. Biotechnology Bulletin, 2023, 39(10): 50-57. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||