








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (9): 281-290.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0085
Previous Articles Next Articles
LIU Hao( ), MA Shi-jie, ZHOU Zhe-min, CUI Wen-jing(
), MA Shi-jie, ZHOU Zhe-min, CUI Wen-jing( )
)
Received:2023-02-07
Online:2023-09-26
Published:2023-10-24
Contact:
CUI Wen-jing
E-mail:6200208115@stu.jiangnan.edu.cn;wjcui@jiangnan.edu.cn
LIU Hao, MA Shi-jie, ZHOU Zhe-min, CUI Wen-jing. Improving the Activity of L-aspartate-a-decarboxylase from Corynebacterium jeikeium Through Semi-rational Design and Whole-cell Catalytic Synthesis of β-alanine[J]. Biotechnology Bulletin, 2023, 39(9): 281-290.
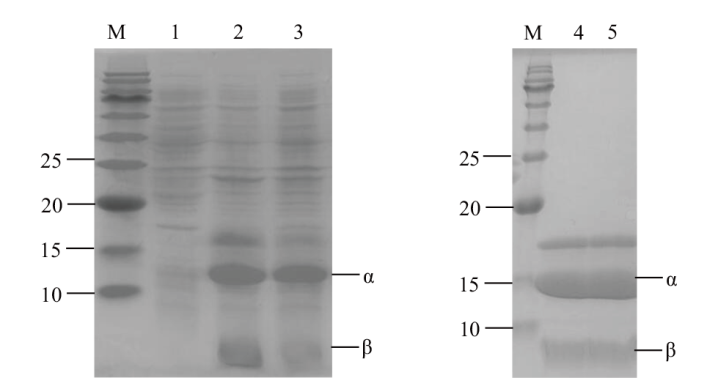
Fig. 1 SDS-PAGE analysis of recombinant enzyme CJpanD and mutants M: Protein marker; 1: E. coli BL21/pET28a cell breaking supernatant; 2: E. coli BL21/pET28a-CJPanD cell breaking supernatant; 3: E. coli BL21/pET28a-CJPanD-L39A cell breaking supernatant; 4: purified pET28a-CJPanD; 5: purified pET28a-CJPanD-L39A
| Mutant | ddG/(Kal·mol-1) |
|---|---|
| H11 | -3.572 |
| C26 | -4.946 |
| L39 | -3.137 |
| I49 | -1.518 |
| N72 | -3.458 |
| L55 | -2.940 |
| T56 | -2.283 |
Table 1 Selection of pseudo-mutant amino acids
| Mutant | ddG/(Kal·mol-1) |
|---|---|
| H11 | -3.572 |
| C26 | -4.946 |
| L39 | -3.137 |
| I49 | -1.518 |
| N72 | -3.458 |
| L55 | -2.940 |
| T56 | -2.283 |
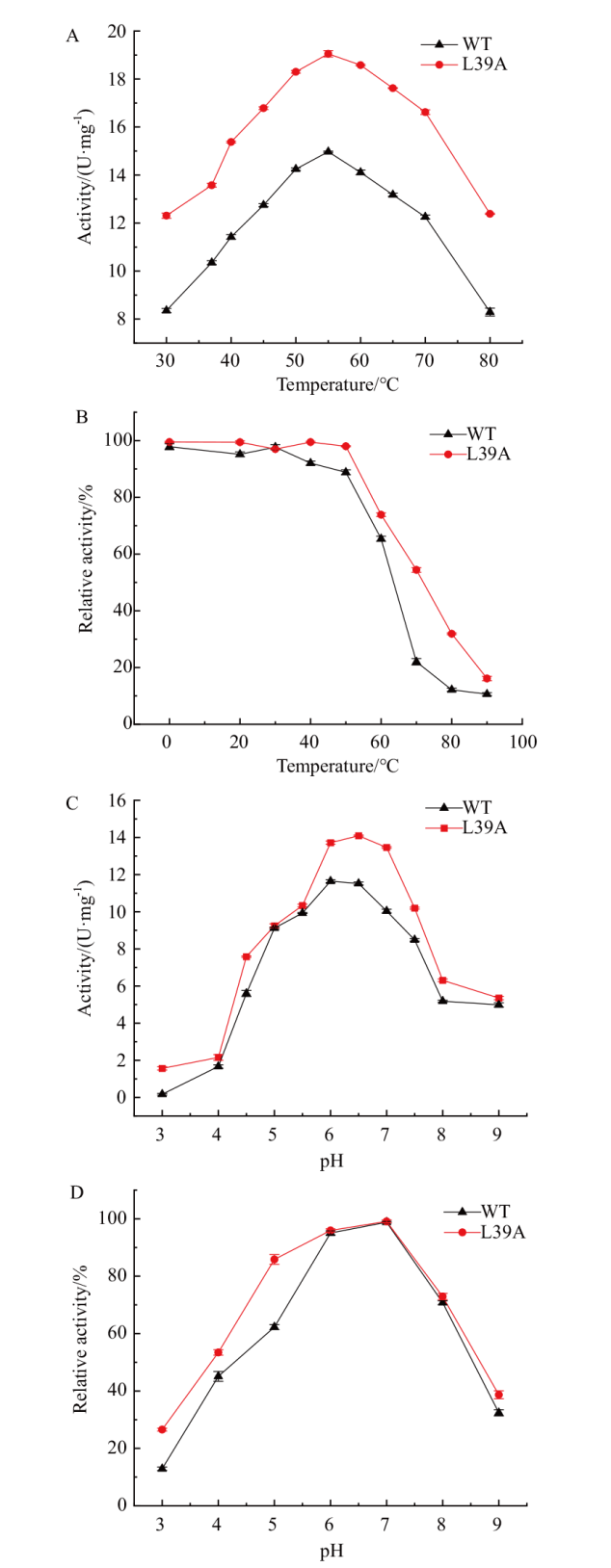
Fig. 4 Optimal temperature and temperature stability, optimal pH and pH stability of recombinant enzymes A: Optimal temperature for recombinant enzymes. B: Effect of temperature on the stability of recombinant enzymes. C: Optimal pH for recombinant enzymes. D: Effect of pH on the stability of recombinant enzymes
| Mutant | Km/ (mmol·L-1) | Kcat/ (s-1) | Kcat/Km / (mmol·L-1·s-1) |
|---|---|---|---|
| WT | 3.6±0.1 | 102.1±0.3 | 28.1 |
| L39A | 3.3±0.1 | 132.5±0.3 | 39.3 |
Table 2 Kinetic parameters of WT and L39A
| Mutant | Km/ (mmol·L-1) | Kcat/ (s-1) | Kcat/Km / (mmol·L-1·s-1) |
|---|---|---|---|
| WT | 3.6±0.1 | 102.1±0.3 | 28.1 |
| L39A | 3.3±0.1 | 132.5±0.3 | 39.3 |
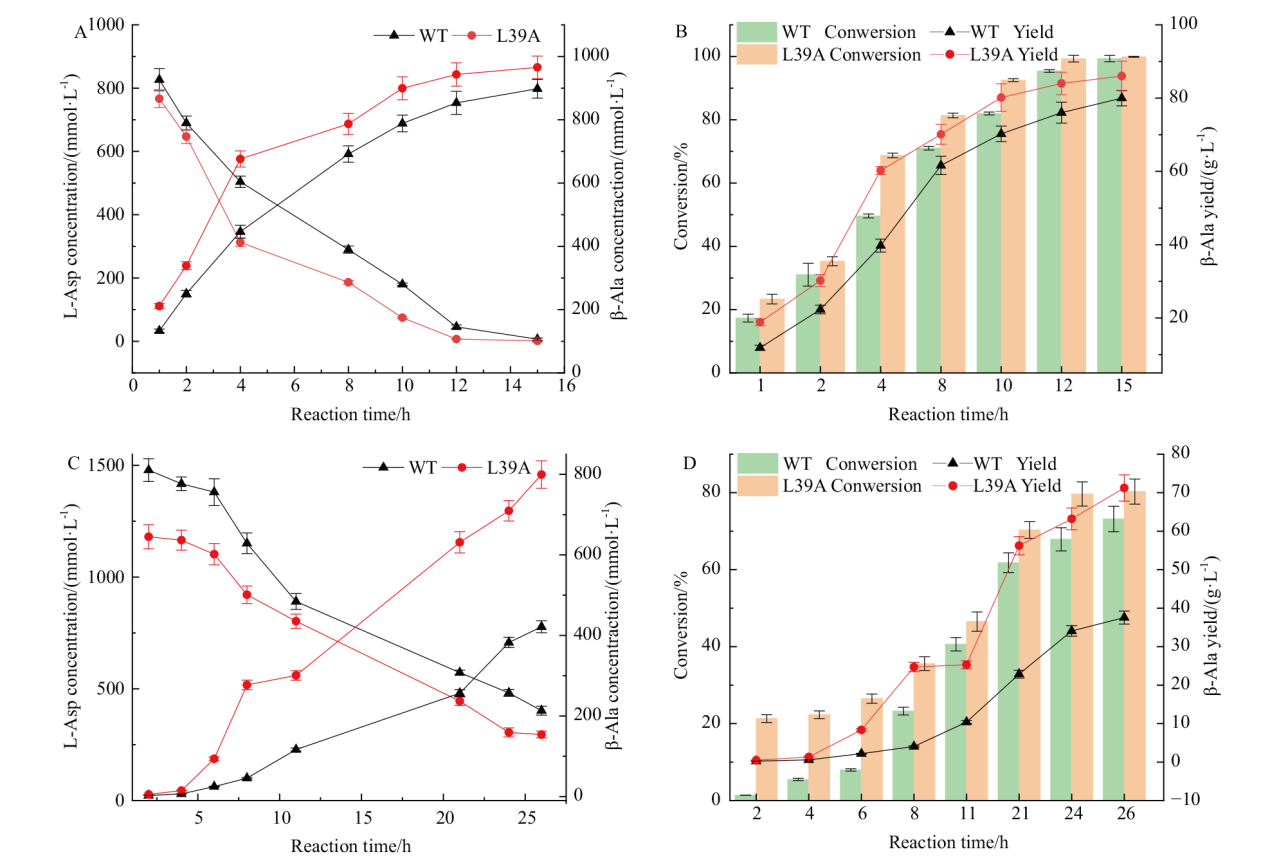
Fig. 7 Whole cell catalytic synthesis of β-alanine A: 1 mol/L substrate, change in substrate and product. B: 1 mol/L substrate, change of conversion and yield. C: 1.5 mol/L substrate, change of substrate and product. D: 1.5 mol/L substrate, change of conversion and yield
| 26 h | Conversion/% | β-alanine yield/(g·L-1) |
|---|---|---|
| WT | 73.1 | 37.55 |
| L39A | 80.2 | 71.22 |
Table 3 Conversion and yield at 26 h per batch
| 26 h | Conversion/% | β-alanine yield/(g·L-1) |
|---|---|---|
| WT | 73.1 | 37.55 |
| L39A | 80.2 | 71.22 |
| [1] |
Brencher L, Verhaegh R, Kirsch M. Attenuation of intestinal ischemia-reperfusion-injury by β-alanine: a potentially glycine-receptor mediated effect[J]. J Surg Res, 2017, 211: 233-241.
doi: S0022-4804(16)30573-X pmid: 28501123 |
| [2] |
Blancquaert L, Baba SP, Kwiatkowski S, et al. Carnosine and anserine homeostasis in skeletal muscle and heart is controlled by β-alanine transamination[J]. J Physiol, 2016, 594(17): 4849-4863.
doi: 10.1113/tjp.2016.594.issue-17 URL |
| [3] |
Quesnele J, Laframboise M, Wong J, et al. The effects of beta-alanine supplementation on performance: a systematic review of the literature[J]. Int J Sport Nutr Exerc Metab, 2013, 24(1): 14-27.
doi: 10.1123/ijsnem.2013-0007 URL |
| [4] |
Dusch N, Pühler A, Kalinowski J. Expression of the Corynebacterium glutamicum panD gene encoding L-aspartate-α-decarboxylase leads to pantothenate overproduction in Escherichia coli[J]. Appl Environ Microbiol, 1999, 65(4): 1530-1539.
doi: 10.1128/AEM.65.4.1530-1539.1999 URL |
| [5] |
Jang YS, Kim B, Shin JH, et al. Bio-based production of C2-C6 platform chemicals[J]. Biotechnol Bioeng, 2012, 109(10): 2437-2459.
doi: 10.1002/bit.v109.10 URL |
| [6] |
Williamson JM, Brown GM. Purification and properties of L-Aspartate-alpha-decarboxylase, an enzyme that catalyzes the formation of beta-alanine in Escherichia coli[J]. J Biol Chem, 1979, 254(16): 8074-8082.
doi: 10.1016/S0021-9258(18)36052-6 URL |
| [7] |
Chopra S, Pai H, Ranganathan A. Expression,purification,and biochemical characterization of Mycobacterium tuberculosis aspartate decarboxylase,PanD[J]. Protein Expr. Purif., 2002, 25(3): 533-540.
doi: 10.1016/S1046-5928(02)00039-6 URL |
| [8] | 范海洋. 重组大肠杆菌L-天冬氨酸-α-脱羧酶的制备及应用研究[D]. 上海: 华东理工大学, 2013. |
| Fan HY. Preparation and application of recombinant Escherichia coli L- aspartic acid-α-decarboxylase[D]. Shanghai: East China University of Science and Technology, 2013. | |
| [9] | 赵连真, 张梁, 石贵阳. 谷氨酸棒杆菌L-天冬氨酸α-脱羧酶在大肠杆菌中的表达及酶转化生产β-丙氨酸[J]. 微生物学通报, 2013, 40(12): 2161-2170. |
| Zhao LZ, Zhang L, Shi G. Expression of L-aspartate α-decarboxylase from Corynebacterium glutamicum in Escherichia coli and its application in enzymatic synthesis of β-alanine[J]. Microbiol China, 2013, 40(12): 2161-2170. | |
| [10] | 邓思颖, 张君丽, 蔡真, 等. 枯草芽胞杆菌L-天冬氨酸α脱羧酶的酶学性质[J]. 生物工程学报, 2015, 31(8): 1184-1193. |
| Deng SY, Zhang JL, Cai Z, et al. Characterization of L-aspartate-α-decarboxylase from Bacillus subtilis[J]. Chin J Biotechnol, 2015, 31(8): 1184-1193. | |
| [11] | 陈涛, 徐世永, 冯炎. 结核杆菌L 天冬氨酸α脱羧酶诱导表达条件研究[J]. 金陵科技学院学报, 2016, 32(3)80-83. |
| Chen T, Xu SY, Feng Y. Inducing conditions of recombined L-aspartate-α-decarboxylase in fermentor[J]. J Jinling Inst Technol, 2016, 32(3)80-83. | |
| [12] |
Ran KA, Il LB, Woo HB, et al. Crystallization and preliminary X-ray crystallographic analysis of aspartate 1-decarboxylase from Helicobacter pylori[J]. Acta Crystallogr Sect D Biol Crystallogr, 2002, 58(Pt 5): 861-3.
doi: 10.1107/S0907444902004833 URL |
| [13] |
Yu XJ, Huang CY, Xu XD, et al. Protein engineering of a pyridoxal-5'-phosphate-dependent l-aspartate-α-decarboxylase from Tribolium castaneum for β-alanine production[J]. Molecules, 2020, 25(6): 1280.
doi: 10.3390/molecules25061280 URL |
| [14] |
Monteiro DF, Patel V, Bartlett C, et al. The structure of the PanD/PanZ protein complex reveals negative feedback regulation of pantothenate biosynthesis by coenzyme A[J]. Chem Biol, 2015, 22(4): 492-503.
doi: 10.1016/j.chembiol.2015.03.017 URL |
| [15] |
Fouad WM, Rathinasabapathi B. Expression of bacterial l-aspartate-α-decarboxylase in tobacco increases β-alanine and pantothenate levels and improves thermotolerance[J]. Plant Mol Biol, 2006, 60(4): 495-505.
doi: 10.1007/s11103-005-4844-9 URL |
| [16] | 张腾辉. L-天冬氨酸α-脱羧酶的表达、改造及全细胞制备β-丙氨酸[D]. 无锡: 江南大学, 2018. |
| Zhang TH. Expression and transformation of L- aspartate α-decarboxylase and preparation of β-alanine by whole cell[D]. Wuxi: Jiangnan University, 2018. | |
| [17] | 张潇潇. 钝齿棒杆菌L-天冬氨酸α-脱羧酶基因的克隆与表达[D]. 杭州: 浙江工业大学, 2008. |
| Zhang XX. Cloning and expression of l-aspartate α-decarboxylase gene from Corynebacterium crenatum[D]. Hangzhou: Zhejiang University of Technology, 2008. | |
| [18] |
范雪萍, 冯志彬, 房美芳, 等. 特基拉芽孢杆菌L-天冬氨酸α-脱羧酶的异源表达及高密度发酵[J]. 食品科学, 2018, 39(2): 144-150.
doi: 10.7506/spkx1002-6630-201802023 |
| Fan XP, Feng ZB, Fang MF, et al. Heterologous expression of the Bacillus tequilensis L-aspartate α-decarboxylase in Escherichia coli and its high cell density fermentation[J]. Food Sci, 2018, 39(2): 144-150. | |
| [19] | Li HH, Lu XL, Chen KQ, et al. β-alanine production using whole-cell biocatalysts in recombinant Escherichia coli[J]. Mol Catal, 2018, 449: 93-98. |
| [20] |
Zhang TH, Zhang RZ, Xu MJ, et al. Glu56Ser mutation improves the enzymatic activity and catalytic stability of Bacillus subtilis l-aspartate α-decarboxylase for an efficient β-alanine production[J]. Process Biochem, 2018, 70: 117-123.
doi: 10.1016/j.procbio.2018.04.004 URL |
| [21] |
Pei WL, Zhang JL, Deng SY, et al. Molecular engineering of l-aspartate-α-decarboxylase for improved activity and catalytic stability[J]. Appl Microbiol Biotechnol, 2017, 101(15): 6015-6021.
doi: 10.1007/s00253-017-8337-y pmid: 28589224 |
| [22] | 陈虹. L-天冬氨酸-α-脱羧酶的蛋白质工程改造及其在β-丙氨酸生产中的应用[D]. 杭州: 浙江工业大学, 2019. |
| Chen H. Engineering transformation of L- aspartate-α-decarboxylase in protein and its application in the production of β-alanine[D]. Hangzhou: Zhejiang University of Technology, 2019. | |
| [23] |
Xu JZ, Yang HK, Zhang WG. NADPH metabolism: a survey of its theoretical characteristics and manipulation strategies in amino acid biosynthesis[J]. Crit Rev Biotechnol, 2018, 38(7): 1061-1076.
doi: 10.1080/07388551.2018.1437387 URL |
| [24] | Qian YY, Liu J, Song W, et al. Production of β-alanine from fumaric acid using a dual-enzyme cascade[J]. Chem Cat Chem, 2018, 10(21): 4984-4991. |
| [25] |
Borodina I, Kildegaard KR, Jensen NB, et al. Establishing a synthetic pathway for high-level production of 3-hydroxypropionic acid in Saccharomyces cerevisiae via β-alanine[J]. Metab Eng, 2015, 27: 57-64.
doi: S1096-7176(14)00125-6 pmid: 25447643 |
| [26] |
Wang L, Piao XY, Cui SM, et al. Enhanced production of β-alanine through co-expressing two different subtypes of L-aspartate-α-decarboxylase[J]. J Ind Microbiol Biotechnol, 2020, 47(6): 465-474.
doi: 10.1007/s10295-020-02285-5 URL |
| [27] |
Sharma R, Kothapalli R, Van Dongen AMJ, et al. Chemoinformatic identification of novel inhibitors against Mycobacterium tuberculosis L-aspartate α-decarboxylase[J]. PLoS One, 2012, 7(3): e33521.
doi: 10.1371/journal.pone.0033521 URL |
| [28] | 莫芹, 李由然, 石贵阳. 细菌L-天冬氨酸α-脱羧酶的分子机制及分子改造研究进展[J]. 微生物学通报, 2018, 45(7): 1546-1554. |
| Mo Q, Li YR, Shi GY. Advances in molecular mechanism and modification of bacterial L-aspartate alpha-decarboxylase[J]. Microbiol China, 2018, 45(7): 1546-1554. | |
| [29] |
Parker JMR, Hodges RS. HomologyPlot: Searching for homology to a family of proteins using a database of unique conserved patterns[J]. J Computer-Aided Mol Des, 1994, 8(2): 193-210.
doi: 10.1007/BF00119867 URL |
| [30] |
Sun ZT, Lonsdale R, Wu L, et al. Structure-guided triple-code saturation mutagenesis: efficient tuning of the stereoselectivity of an epoxide hydrolase[J]. ACS Catal, 2016, 6(3): 1590-1597.
doi: 10.1021/acscatal.5b02751 URL |
| [31] |
Denard CA, Ren HQ, Zhao HM. Improving and repurposing biocatalysts via directed evolution[J]. Curr Opin Chem Biol, 2015, 25: 55-64.
doi: 10.1016/j.cbpa.2014.12.036 pmid: 25579451 |
| [32] |
Sun ZT, Wikmark Y, Bäckvall JE, et al. New concepts for increasing the efficiency in directed evolution of stereoselective enzymes[J]. Chem - A Eur J, 2016, 22(15): 5046-5054.
doi: 10.1002/chem.v22.15 URL |
| [33] |
Dehouck Y, Grosfils A, Folch B, et al. Fast and accurate predictions of protein stability changes upon mutations using statistical potentials and neural networks[J]. Bioinformatics, 2009, 25(19): 2537-2543.
doi: 10.1093/bioinformatics/btp445 pmid: 19654118 |
| [34] |
Kumar V, Rahman S, Choudhry H, et al. Computing disease-linked SOD1 mutations: deciphering protein stability and patient-phenotype relations[J]. Sci Rep, 2017, 7(1): 4678.
doi: 10.1038/s41598-017-04950-9 pmid: 28680046 |
| [35] |
Jumper J, Evans R, Pritzel A, et al. Highly accurate protein structure prediction with AlphaFold[J]. Nature, 2021, 596(7873): 583-589.
doi: 10.1038/s41586-021-03819-2 |
| [36] |
Jiménez-Osés G, Osuna S, Gao X, et al. The role of distant mutations and allosteric regulation on LovD active site dynamics[J]. Nat Chem Biol, 2014, 10(6): 431-436.
doi: 10.1038/nchembio.1503 pmid: 24727900 |
| No related articles found! |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||