








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (2): 197-211.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0830
Previous Articles Next Articles
XIN Qi( ), LI Ya-fan, YIN Zheng, ZHANG Xiao-dan, CHEN Ting, LIU Xiao-hua(
), LI Ya-fan, YIN Zheng, ZHANG Xiao-dan, CHEN Ting, LIU Xiao-hua( )
)
Received:2023-08-25
Online:2024-02-26
Published:2024-03-13
Contact:
LIU Xiao-hua
E-mail:704427611@qq.com;3305@ldu.edu.cn
XIN Qi, LI Ya-fan, YIN Zheng, ZHANG Xiao-dan, CHEN Ting, LIU Xiao-hua. Identification and Expression Analysis of the CBL-CIPK Gene Family in Sugarcane[J]. Biotechnology Bulletin, 2024, 40(2): 197-211.
| 基因 Gene | 基因ID Gene ID | 基因长度 Gene length /bp | 正负链 Strand | mRNA长度 mRNA length/bp | 氨基酸数目 Amino acid/aa | 等电点 pI | 外显子个数 Exon number | 染色体定位 Chromosome | 相对分子质量 Molecular weight/kD | 亚细胞定位预测 Subcellular localization |
|---|---|---|---|---|---|---|---|---|---|---|
| SsCBL1 | Sspon.01G0014730-1A | 3 124 | + | 579 | 193 | 5.00 | 7 | Chr.1A | 22.26 | Cyto |
| SsCBL2 | Sspon.01G0023690-1A | 3 641 | + | 675 | 225 | 4.82 | 8 | Chr.1A | 25.77 | Cyto |
| SsCBL3 | Sspon.01G0014730-2B | 3 431 | - | 639 | 213 | 4.75 | 8 | Chr.1B | 24.49 | Cyto |
| SsCBL4 | Sspon.01G0014730-3C | 3 431 | - | 639 | 213 | 4.75 | 8 | Chr.1C | 24.49 | Cyto |
| SsCBL5 | Sspon.01G0014730-4D | 3 431 | - | 639 | 213 | 4.75 | 8 | Chr.1D | 24.49 | Cyto |
| SsCBL6 | Sspon.01G0023690-3D | 3 641 | + | 675 | 225 | 4.82 | 8 | Chr.1D | 25.77 | Cyto |
| SsCBL7 | Sspon.02G0030420-1A | 3 582 | + | 660 | 220 | 4.92 | 9 | Chr.2A | 25.43 | Chlo |
| SsCBL8 | Sspon.02G0030420-2C | 3 640 | - | 675 | 225 | 4.79 | 8 | Chr.2C | 25.84 | Cyto |
| SsCBL9 | Sspon.03G0011980-1A | 4 077 | - | 738 | 246 | 6.14 | 10 | Chr.3A | 28.83 | Cyto |
| SsCBL10 | Sspon.03G0012700-1A | 4 617 | - | 882 | 294 | 4.84 | 9 | Chr.3A | 33.11 | E.R. |
| SsCBL11 | Sspon.03G0031480-1B | 4 573 | - | 864 | 288 | 4.84 | 8 | Chr.3B | 32.40 | Chlo |
| SsCBL12 | Sspon.03G0011980-2B | 4 758 | + | 882 | 294 | 5.28 | 10 | Chr.3B | 33.94 | Cyto |
| SsCBL13 | Sspon.03G0011980-1P | 3 620 | - | 660 | 220 | 6.19 | 8 | Chr.3B | 25.66 | Cyto |
| SsCBL14 | Sspon.04G0013450-2B | 3 432 | + | 639 | 213 | 4.77 | 8 | Chr.4B | 24.34 | Nucl |
| SsCBL15 | Sspon.07G0003410-1A | 3 363 | - | 633 | 211 | 4.88 | 8 | Chr.7A | 24.11 | Chlo |
| SsCBL16 | Sspon.07G0018960-1A | 3 619 | - | 669 | 223 | 4.82 | 8 | Chr.7A | 25.68 | Chlo |
| SsCBL17 | Sspon.07G0018960-2B | 3 363 | - | 618 | 206 | 4.99 | 7 | Chr.7B | 23.77 | Chlo |
| SsCBL18 | Sspon.07G0018960-3C | 3 324 | + | 612 | 204 | 4.91 | 7 | Chr.7C | 23.52 | Mito |
| SsCBL19 | Sspon.07G0018960-4D | 3 485 | + | 642 | 214 | 4.92 | 8 | Chr.7D | 24.69 | Chlo |
Table 1 Statistical table of physico-chemical properties of 19 CBLs of sugarcane
| 基因 Gene | 基因ID Gene ID | 基因长度 Gene length /bp | 正负链 Strand | mRNA长度 mRNA length/bp | 氨基酸数目 Amino acid/aa | 等电点 pI | 外显子个数 Exon number | 染色体定位 Chromosome | 相对分子质量 Molecular weight/kD | 亚细胞定位预测 Subcellular localization |
|---|---|---|---|---|---|---|---|---|---|---|
| SsCBL1 | Sspon.01G0014730-1A | 3 124 | + | 579 | 193 | 5.00 | 7 | Chr.1A | 22.26 | Cyto |
| SsCBL2 | Sspon.01G0023690-1A | 3 641 | + | 675 | 225 | 4.82 | 8 | Chr.1A | 25.77 | Cyto |
| SsCBL3 | Sspon.01G0014730-2B | 3 431 | - | 639 | 213 | 4.75 | 8 | Chr.1B | 24.49 | Cyto |
| SsCBL4 | Sspon.01G0014730-3C | 3 431 | - | 639 | 213 | 4.75 | 8 | Chr.1C | 24.49 | Cyto |
| SsCBL5 | Sspon.01G0014730-4D | 3 431 | - | 639 | 213 | 4.75 | 8 | Chr.1D | 24.49 | Cyto |
| SsCBL6 | Sspon.01G0023690-3D | 3 641 | + | 675 | 225 | 4.82 | 8 | Chr.1D | 25.77 | Cyto |
| SsCBL7 | Sspon.02G0030420-1A | 3 582 | + | 660 | 220 | 4.92 | 9 | Chr.2A | 25.43 | Chlo |
| SsCBL8 | Sspon.02G0030420-2C | 3 640 | - | 675 | 225 | 4.79 | 8 | Chr.2C | 25.84 | Cyto |
| SsCBL9 | Sspon.03G0011980-1A | 4 077 | - | 738 | 246 | 6.14 | 10 | Chr.3A | 28.83 | Cyto |
| SsCBL10 | Sspon.03G0012700-1A | 4 617 | - | 882 | 294 | 4.84 | 9 | Chr.3A | 33.11 | E.R. |
| SsCBL11 | Sspon.03G0031480-1B | 4 573 | - | 864 | 288 | 4.84 | 8 | Chr.3B | 32.40 | Chlo |
| SsCBL12 | Sspon.03G0011980-2B | 4 758 | + | 882 | 294 | 5.28 | 10 | Chr.3B | 33.94 | Cyto |
| SsCBL13 | Sspon.03G0011980-1P | 3 620 | - | 660 | 220 | 6.19 | 8 | Chr.3B | 25.66 | Cyto |
| SsCBL14 | Sspon.04G0013450-2B | 3 432 | + | 639 | 213 | 4.77 | 8 | Chr.4B | 24.34 | Nucl |
| SsCBL15 | Sspon.07G0003410-1A | 3 363 | - | 633 | 211 | 4.88 | 8 | Chr.7A | 24.11 | Chlo |
| SsCBL16 | Sspon.07G0018960-1A | 3 619 | - | 669 | 223 | 4.82 | 8 | Chr.7A | 25.68 | Chlo |
| SsCBL17 | Sspon.07G0018960-2B | 3 363 | - | 618 | 206 | 4.99 | 7 | Chr.7B | 23.77 | Chlo |
| SsCBL18 | Sspon.07G0018960-3C | 3 324 | + | 612 | 204 | 4.91 | 7 | Chr.7C | 23.52 | Mito |
| SsCBL19 | Sspon.07G0018960-4D | 3 485 | + | 642 | 214 | 4.92 | 8 | Chr.7D | 24.69 | Chlo |

Fig. 1 Domain and motif analysis of CBL-CIPK gene family A: Analysis of motif, domain of sugarcane CBL family members. B: Analysis of motif, domain of sugarcane CIPK family members. From left to right are the evolutionary relationship tree, motif and domain, with different colored modules representing different motifs and domains

Fig. 2 Analysis in the gene structures of CBL-CIPK gene family A: Gene structure of sugarcane CBL family members. B: Gene structure of sugarcane CIPK family members. Evolutionary relationship tree on the left, gene structure of family members on the right, UTR in green and CDS in yellow
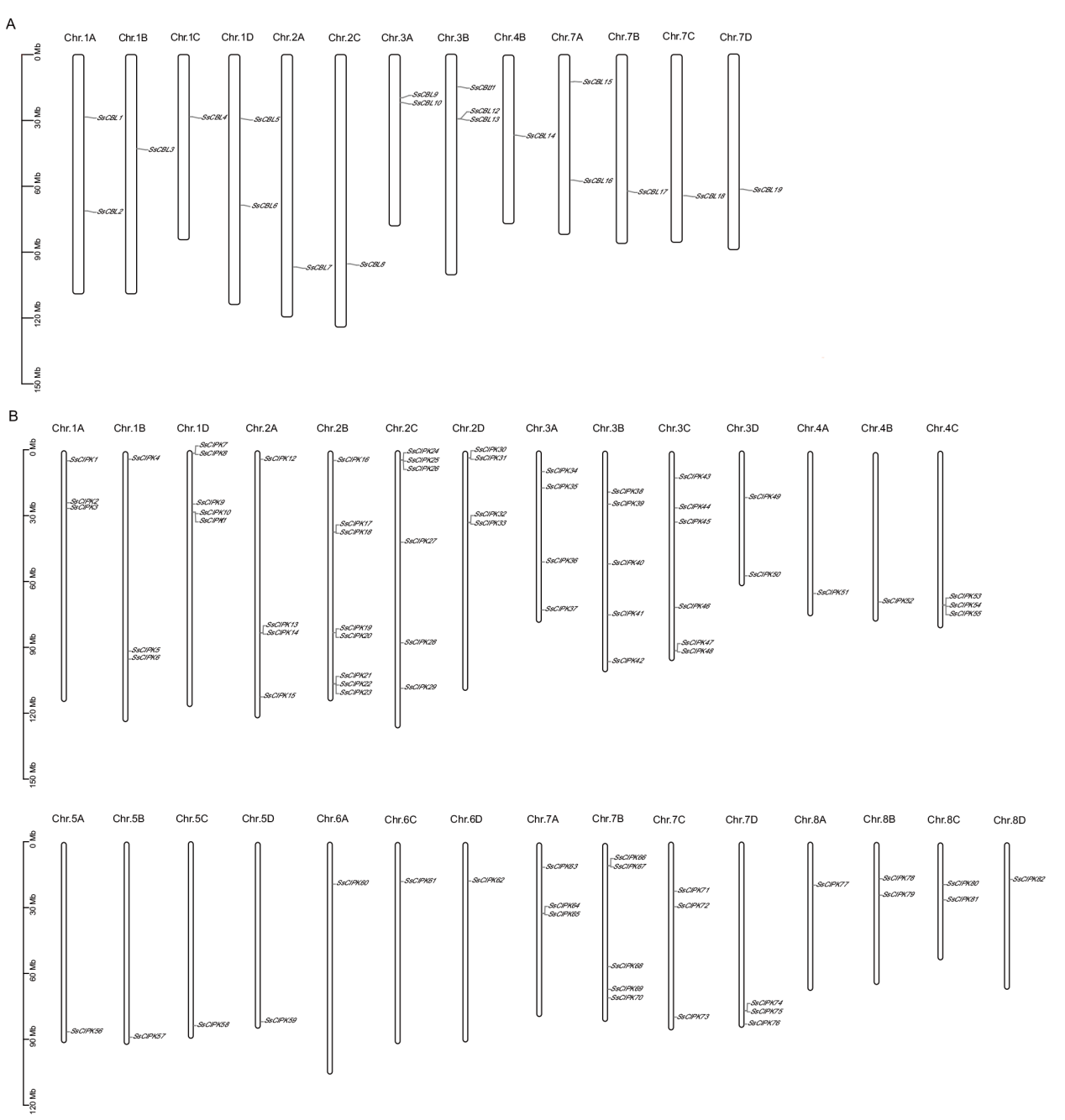
Fig. 3 Analysis for their positions on the chromosomes in CBL-CIPK gene family A: Chromosomal localization of sugarcane CBL family members. B: Chromosomal localization of sugarcane CIPK family members
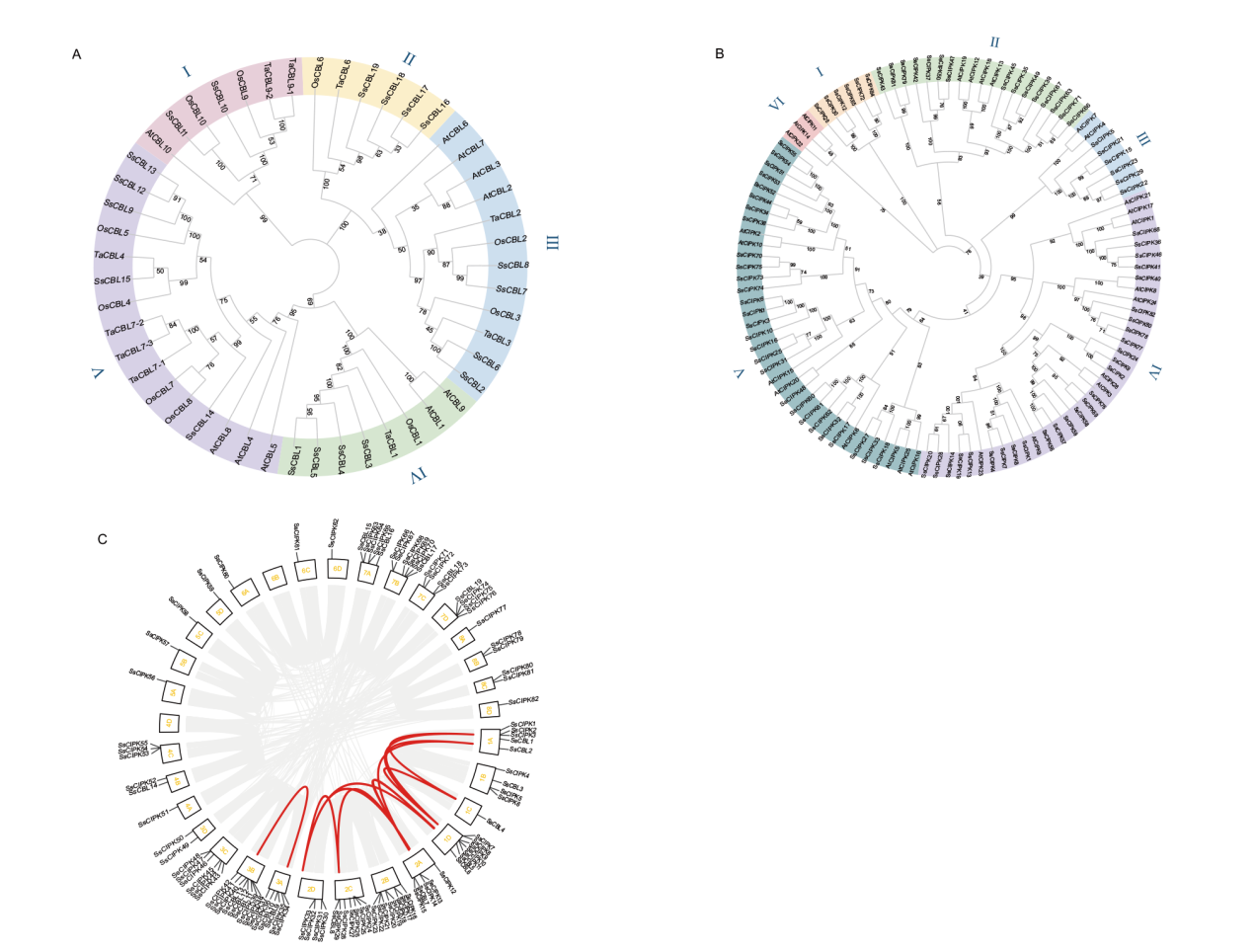
Fig. 5 Phylogenetic tree and covariance analysis of the CBL-CIPK gene family A: Phylogenetic evolutionary tree of the multispecies CBL family. B: Phylogenetic evolutionary tree of the multispecies CIPK family. C: Analysis of intraspecific CBL-CIPK gene family covariance in sugarcane. All gene pairs in the whole genome are gray, CBL-CIPK family members form gene pairs in red, sugarcane is a heterotetraploid, and 1A-8D are the 32 chromosomes of sugarcane

Fig. 6 RT-qPCR analysis of SsCBLs, SsCIPKs genes involved in different temperature stresses response A: Gene expression profiles of SsCBLs under 4℃ treatment. B: Gene expression profiles of SsCIPKs under 4℃ treatment. C: Gene expression profiles of SsCBLs under 45℃ treatment. D: Gene expression profiles of SsCIPKs under 45℃ treatment

Fig. 7 RT-qPCR analysis of SsCBLs, SsCIPKs genes involved in NaHCO3/PEG stresses A: Gene expression profiles of SsCBLs under170 mmol/L NaHCO3 treatment. B: Gene expression profiles of SsCIPKs under 170 mmol/L NaHCO3 treatment. C: Gene expression profiles of SsCBLs under 150 g/L PEG-6000 treatment. D: Gene expression profiles of SsCIPKs under 150 g/L PEG-6000 treatment
| [1] |
Plasencia FA, Estrada Y, Flores FB, et al. The Ca2+ sensor calcineurin B-like protein 10 in plants: Emerging new crucial roles for plant abiotic stress tolerance[J]. Front Plant Sci, 2021, 11: 599944.
doi: 10.3389/fpls.2020.599944 URL |
| [2] | Niu L, Dong B, Song Z, et al. Genome-wide identification and characterization of CIPK family and analysis responses to various stresses in apple(Malus domestica)[J]. Int J Mol Sci, 2018, 19(7): E2131. |
| [3] |
Hashimoto K, Eckert C, Anschütz U, et al. Phosphorylation of calcineurin B-like(CBL)calcium sensor proteins by their CBL-interacting protein kinases(CIPKs)is required for full activity of CBL-CIPK complexes toward their target proteins[J]. J Biol Chem, 2012, 287(11): 7956-7968.
doi: 10.1074/jbc.M111.279331 pmid: 22253446 |
| [4] | Chaves-Sanjuan A, Sanchez-Barrena MJ, Gonzalez-Rubio JM, et al. Structural basis of the regulatory mechanism of the plant CIPK family of protein kinases controlling ion homeostasis and abiotic stress[J]. Proc Natl Acad Sci USA, 2014, 111(42): E4532-E4541. |
| [5] |
Ma X, Li QH, Yu YN, et al. The CBL-CIPK pathway in plant response to stress signals[J]. Int J Mol Sci, 2020, 21(16): 5668.
doi: 10.3390/ijms21165668 URL |
| [6] |
Mo C, Wan S, Xia Y, et al. Expression patterns and identified protein-protein interactions suggest that cassava CBL-CIPK signal networks function in responses to abiotic stresses[J]. Front Plant Sci, 2018, 9: 269.
doi: 10.3389/fpls.2018.00269 pmid: 29552024 |
| [7] |
Sánchez-Barrena M, Martínez-Ripoll M, Albert A. Structural biology of a major signaling network that regulates plant abiotic stress: The CBL-CIPK mediated pathway[J]. Int J Mol Sci, 2013, 14(3): 5734-5749.
doi: 10.3390/ijms14035734 pmid: 23481636 |
| [8] |
Poddar N, Deepika D, Chitkara P, et al. Molecular and expression analysis indicate the role of CBL interacting protein kinases(CIPKs)in abiotic stress signaling and development in chickpea[J]. Sci Rep, 2022, 12(1): 16862.
doi: 10.1038/s41598-022-20750-2 |
| [9] |
Ródenas R, Vert G. Regulation of root nutrient transporters by CIPK23: “one kinase to rule them all”[J]. Plant Cell Physiol, 2021, 62(4): 553-563.
doi: 10.1093/pcp/pcaa156 pmid: 33367898 |
| [10] |
Yin X, Xia Y, Xie Q, et al. The protein kinase complex CBL10-CIPK8-SOS1 functions in Arabidopsis to regulate salt tolerance[J]. J Exp Bot, 2020, 71(6): 1801-1814.
doi: 10.1093/jxb/erz549 URL |
| [11] |
Imtiaz K, Ahmed M, Annum N, et al. AtCIPK16, a CBL-interacting protein kinase gene, confers salinity tolerance in transgenic wheat[J]. Front Plant Sci, 2023, 14: 1127311.
doi: 10.3389/fpls.2023.1127311 URL |
| [12] |
Gu S, Abid M, Bai D, et al. Transcriptome-wide identification and functional characterization of CIPK gene family members in actinidia valvata under salt stress[J]. Int J Mol Sci, 2023, 24(1): 805.
doi: 10.3390/ijms24010805 URL |
| [13] |
Chen L, Wang QQ, Zhou L, et al. Arabidopsis CBL-interacting protein kinase(CIPK6)is involved in plant response to salt/osmotic stress and ABA[J]. Mol Biol Rep, 2013, 40(8): 4759-4767.
doi: 10.1007/s11033-013-2572-9 URL |
| [14] |
Cui XY, Du YT, Fu JD, et al. Wheat CBL-interacting protein kinase 23 positively regulates drought stress and ABA responses[J]. BMC Plant Biol, 2018, 18(1): 93.
doi: 10.1186/s12870-018-1306-5 |
| [15] |
Gao C, Lu S, Zhou R, et al. The OsCBL8-OsCIPK17 module regulates seedling growth and confers resistance to heat and drought in rice[J]. Int J Mol Sci, 2022, 23(20): 12451.
doi: 10.3390/ijms232012451 URL |
| [16] |
Meng D, Dong BY, Niu LL, et al. The pigeon pea CcCIPK14-CcCBL1 pair positively modulates drought tolerance by enhancing flavonoid biosynthesis[J]. Plant J, 2021, 106(5): 1278-1297.
doi: 10.1111/tpj.v106.5 URL |
| [17] |
Yang L, Zhang N, Wang K, et al. CBL-interacting protein kinases 18(CIPK18)gene positively regulates drought resistance in potato[J]. Int J Mol Sci, 2023, 24(4): 3613.
doi: 10.3390/ijms24043613 URL |
| [18] |
Yang S, Li J, Lu J, et al. Potato calcineurin B-like protein CBL4, interacting with calcineurin B-like protein-interacting protein kinase CIPK2, positively regulates plant resistance to stem canker caused by Rhizoctonia solani[J]. Front Microbiol, 2023, 13: 1032900.
doi: 10.3389/fmicb.2022.1032900 URL |
| [19] |
Piao HL, Xuan YH, Park SH, et al. OsCIPK31, a CBL-interacting protein kinase is involved in germination and seedling growth under abiotic stress conditions in rice plants[J]. Mol Cells, 2010, 30(1): 19-27.
doi: 10.1007/s10059-010-0084-1 URL |
| [20] |
Liu P, Duan YH, Liu C, et al. The calcium sensor TaCBL4 and its interacting protein TaCIPK5 are required for wheat resistance to stripe rust fungus[J]. J Exp Bot, 2018, 69(18): 4443-4457.
doi: 10.1093/jxb/ery227 pmid: 29931351 |
| [21] |
Xi Y, Liu JY, Dong C, et al. The CBL and CIPK gene family in grapevine(Vitis vinifera): Genome-wide analysis and expression profiles in response to various abiotic stresses[J]. Front Plant Sci, 2017, 8: 978.
doi: 10.3389/fpls.2017.00978 URL |
| [22] | Aslam M, Fakher B, Jakada BH, et al. Genome-wide identification and expression profiling of CBL-CIPK gene family in pineapple(Ananas comosus)and the role of AcCBL1 in abiotic and biotic stress response[J]. Biomolecules, 2019, 9(7): E293. |
| [23] |
Zhang HF, Yang B, Liu WZ, et al. Identification and characterization of CBL and CIPK gene families in canola(Brassica napus L.)[J]. BMC Plant Biol, 2014, 14(1): 8.
doi: 10.1186/1471-2229-14-8 |
| [24] |
Yin X, Wang QL, Chen Q, et al. Genome-wide identification and functional analysis of the calcineurin B-like protein and calcineurin B-like protein-interacting protein kinase gene families in turnip(Brassica rapa var. rapa)[J]. Front Plant Sci, 2017, 8: 1191.
doi: 10.3389/fpls.2017.01191 URL |
| [25] |
Wang QW, Zhao K, Gong YQ, et al. Genome-wide identification and functional analysis of the calcineurin B-like protein and calcineurin B-like protein-interacting protein kinase gene families in chinese cabbage(Brassica rapa ssp. pekinensis)[J]. Genes, 2022, 13(5): 795.
doi: 10.3390/genes13050795 URL |
| [26] |
Mao JJ, Manik SM, Shi SJ, et al. Mechanisms and physiological roles of the CBL-CIPK networking system in Arabidopsis thaliana[J]. Genes, 2016, 7(9): 62.
doi: 10.3390/genes7090062 URL |
| [27] |
Chen Y, Jin YF, Wang YS, et al. Diverse roles of the CIPK gene family in transcription regulation and various biotic and abiotic stresses: A literature review and bibliometric study[J]. Front Genet, 2022, 13: 1041078.
doi: 10.3389/fgene.2022.1041078 URL |
| [28] |
Zhu XL, Wang BQ, Wang X, et al. Identification of the CIPK-CBL family gene and functional characterization of CqCIPK14 gene under drought stress in quinoa[J]. BMC Genomics, 2022, 23(1): 447.
doi: 10.1186/s12864-022-08683-6 pmid: 35710332 |
| [29] |
Zhang XX, Ren XL, Qi XT, et al. Evolution of the CBL and CIPK gene families in Medicago: genome-wide characterization, pervasive duplication, and expression pattern under salt and drought stress[J]. BMC Plant Biol, 2022, 22(1): 512.
doi: 10.1186/s12870-022-03884-3 |
| [30] |
Tang J, Lin J, Li H, et al. Characterization of CIPK family in Asian pear(Pyrus bretschneideri Rehd)and Co-expression analysis related to salt and osmotic stress responses[J]. Front Plant Sci, 2016, 7: 1361.
doi: 10.3389/fpls.2016.01361 pmid: 27656193 |
| [31] |
Cho JH, Choi MN, Yoon KH, et al. Ectopic expression of sjcbl1, calcineurin b-like 1 gene from sedirea japonica, rescues the salt and osmotic stress hypersensitivity in arabidopsis cbl1 mutant[J]. Front Plant Sci, 2018, 9: 1188.
doi: 10.3389/fpls.2018.01188 URL |
| [32] |
Huang SL, Jiang SF, Liang JS, et al. Roles of plant CBL-CIPK systems in abiotic stress responses[J]. Turk J Bot, 2019, 43(3): 271-280.
doi: 10.3906/bot-1810-35 |
| [33] |
Chao M, Dong J, Hu G, et al. Sequence characteristics and expression analysis of GhCIPK23 gene in upland cotton(Gossypium hirsutum L.)[J]. Int J Mol Sci, 2022, 23(19): 12040.
doi: 10.3390/ijms231912040 URL |
| [34] |
Zhu KK, Chen F, Liu JY, et al. Evolution of an intron-poor cluster of the CIPK gene family and expression in response to drought stress in soybean[J]. Sci Rep, 2016, 6(1): 28225.
doi: 10.1038/srep28225 |
| [35] |
Zhu KK, Fan PH, Liu H, et al. Insight into the CBL and CIPK gene families in pecan(Carya illinoinensis): identification, evolution and expression patterns in drought response[J]. BMC Plant Biol, 2022, 22(1): 221.
doi: 10.1186/s12870-022-03601-0 |
| [36] |
Ma X, Gai WX, Li Y, et al. The CBL-interacting protein kinase CaCIPK13 positively regulates defence mechanisms against cold stress in pepper[J]. J Exp Bot, 2022, 73(5): 1655-1667.
doi: 10.1093/jxb/erab505 pmid: 35137060 |
| [37] |
Xiao C, Zhang H, Xie F, et al. Evolution, gene expression, and protein-protein interaction analyses identify candidate CBL-CIPK signalling networks implicated in stress responses to cold and bacterial infection in citrus[J]. BMC Plant Biol, 2022, 22(1): 420.
doi: 10.1186/s12870-022-03809-0 pmid: 36045357 |
| [38] |
Zhou YL, Cheng Y, Yang YQ, et al. Overexpression of SpCBL6, a calcineurin B-like protein of Stipa purpurea, enhanced cold tolerance and reduced drought tolerance in transgenic Arabidopsis[J]. Mol Biol Rep, 2016, 43(9): 957-966.
doi: 10.1007/s11033-016-4036-5 URL |
| [39] |
Qiu K, Pan H, Sheng Y, et al. The peach(Prunus persica)CBL and CIPK family genes: Protein interaction profiling and expression analysis in response to various abiotic stresses[J]. Plants, 2022, 11(21): 3001.
doi: 10.3390/plants11213001 URL |
| [40] |
Wang S, Li Q. Genome-wide identification of the Salvia miltiorrhiza SmCIPK gene family and revealing the salt resistance characteristic of SmCIPK13[J]. Int J Mol Sci, 2022, 23(12): 6861.
doi: 10.3390/ijms23126861 URL |
| [41] |
Huang L, Li Z, Fu Q, et al. Genome-wide identification of CBL-CIPK gene family in honeysuckle(Lonicera japonica Thunb.)and their regulated expression under salt stress[J]. Front Genet, 2021, 12: 751040.
doi: 10.3389/fgene.2021.751040 URL |
| [42] |
Ma X, Gai WX, Qiao YM, et al. Identification of CBL and CIPK gene families and functional characterization of CaCIPK1 under Phytophthora capsici in pepper(Capsicum annuum L.)[J]. BMC Genomics, 2019, 20(1): 775.
doi: 10.1186/s12864-019-6125-z |
| [43] |
Ma R, Liu W, Li S, et al. Genome-wide identification, characterization and expression analysis of the CIPK gene family in potato(Solanum tuberosum L.)and the role of StCIPK10 in response to drought and osmotic stress[J]. Int J Mol Sci, 2021, 22(24): 13535.
doi: 10.3390/ijms222413535 URL |
| [44] |
Wang L, Feng X, Yao L, et al. Characterization of CBL-CIPK signaling complexes and their involvement in cold response in tea plant[J]. Plant Physiol Biochem, 2020, 154: 195-203.
doi: 10.1016/j.plaphy.2020.06.005 URL |
| [45] |
Sun W, Zhang B, Deng J, et al. Genome-wide analysis of CBL and CIPK family genes in cotton: conserved structures with divergent interactions and expression[J]. Physiol Mol Biol Plants, 2021, 27(2): 359-368.
doi: 10.1007/s12298-021-00943-1 |
| [1] | REN Yan-jing, ZHANG Lu-gang, ZHAO Meng-liang, LI Jiang, SHAO Deng-kui. cDNA Yeast Library Construction of Chinese Cabbage Seeds and Screening and Analysis of BrTTG1 Interacting Proteins [J]. Biotechnology Bulletin, 2024, 40(2): 223-232. |
| [2] | WANG Zi-ying, LONG Chen-jie, FAN Zhao-yu, ZHANG Lei. Screening of OsCRK5-interacted Proteins in Rice Using Yeast Two-hybrid System [J]. Biotechnology Bulletin, 2023, 39(9): 117-125. |
| [3] | WEN Xiao-lei, LI Jian-yuan, LI Na, ZHANG Na, YANG Wen-xiang. Construction and Utilization of Yeast Two-hybrid cDNA Library of Wheat Interacted by Puccinia triticina [J]. Biotechnology Bulletin, 2023, 39(9): 136-146. |
| [4] | ZHAO Xue-ting, GAO Li-yan, WANG Jun-gang, SHEN Qing-qing, ZHANG Shu-zhen, LI Fu-sheng. Cloning and Expression of AP2/ERF Transcription Factor Gene ShERF3 in Sugarcane and Subcellular Localization of Its Encoded Protein [J]. Biotechnology Bulletin, 2023, 39(6): 208-216. |
| [5] | LI Xin-yi, JIANG Chun-xiu, XUE Li, JIANG Hong-tao, YAO Wei, DENG Zu-hu, ZHANG Mu-qing, YU Fan. Enhancing Hybridization Signal of Sugarcane Chromosome Oligonucleotide Probe via Multiple Fluorescence Labeled Primers [J]. Biotechnology Bulletin, 2023, 39(5): 103-111. |
| [6] | HOU Xiao-yuan, CHE Zheng-zheng, LI Heng-jing, DU Chong-yu, XU Qian, WANG Qun-qing. Construction of the Soybean Membrane System cDNA Library and Interaction Proteins Screening for Effector PsAvr3a [J]. Biotechnology Bulletin, 2023, 39(4): 268-276. |
| [7] | LUO Yan-ju, XIE Lin-yan, ZOU Qing-lin, LI Si-jie, LIU Han, LIU Lu-feng, HE Li-lian, LI Fu-sheng. Physiological Response and Drought Resistance Evaluation of Endophytic Bacteria to Sugarcane Under Drought Stress [J]. Biotechnology Bulletin, 2023, 39(12): 219-228. |
| [8] | HUANG Jia-yan, FENG Xiao-yan, SHEN Lin-bo, WANG Wen-zhi, HU Hai-yan, ZHANG Shu-zhen. Cloning of Sugarcane ShPR10 Gene and Study on the Interaction Between ShPR10 Protein and P1 Protein Encoded by Sugarcane Streak Mosaic Virus [J]. Biotechnology Bulletin, 2023, 39(10): 163-174. |
| [9] | WANG Xin-yi, WANG Xiao-qian, WANG Hong-jun, CHAO Yue-hui. Screening, Expression, and Validation of Nanobodies with FLAG Tag [J]. Biotechnology Bulletin, 2023, 39(10): 323-331. |
| [10] | GAO Xiao-ning, LIU Rui, WU Zi-lin, WU Jia-yun. Characteristics of Endophytic Fungal and Bacterial Community in the Stalks of Sugarcane Cultivars Resistant to Ratoon Stunting Disease [J]. Biotechnology Bulletin, 2022, 38(6): 166-173. |
| [11] | ZHANG Jing, YOU Chui-huai, CAO Yue, CUI Tian-zhen, YANG Jing-tao, LUO Jun. Sugarcane Rhizosphere Microecology and Its Relationship with Smut Control [J]. Biotechnology Bulletin, 2022, 38(11): 21-31. |
| [12] | FENG Cui-lian, WAN Yue, WANG Jun-gang, FENG Xiao-yan, ZHAO Ting-ting, WANG Wen-zhi, SHEN Lin-bo, ZHANG Shu-zhen. Establishment of a Transformant-specific Detection Method for Cry1Ac-2A-gna Transgenic Sugarcane BCG-17 [J]. Biotechnology Bulletin, 2021, 37(5): 248-258. |
| [13] | YANG Hua-jie, ZHOU Yu-ping, FAN Tian, LV Tian-xiao, XIE Chu-ping, TIAN Chang-en. Screening and Identification of IQM4-Interacting Proteins in Arabidopsis thaliana [J]. Biotechnology Bulletin, 2021, 37(11): 190-196. |
| [14] | RONG Yu-ping, TANG Bin, LI Peng, ZHANG Jie-qiong, CHEN Qing-fu, ZHU Li-wei, DENG Jiao, HUANG Juan. Identification and Expression of NAC Transcription Factor FtNAC17 in Tartary Buckwheat [J]. Biotechnology Bulletin, 2021, 37(1): 174-181. |
| [15] | FENG Cui-lian, ZHANG Shu-zhen. Breeding of Transgenic Insect-resistant Sugarcane and Strategies for Preventing the Its Resistance to Insects from Loss [J]. Biotechnology Bulletin, 2020, 36(7): 209-219. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||