








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (6): 290-298.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0006
Previous Articles Next Articles
YAN Huan-huan1,2( ), SHANG Yi-tong1,2, WANG Li-hong1,2, TIAN Xue-qin1,2, LIAO Hai-yan1,2, ZENG Bin4, HU Zhi-hong1,2,3(
), SHANG Yi-tong1,2, WANG Li-hong1,2, TIAN Xue-qin1,2, LIAO Hai-yan1,2, ZENG Bin4, HU Zhi-hong1,2,3( )
)
Received:2024-01-04
Online:2024-06-26
Published:2024-05-14
Contact:
HU Zhi-hong
E-mail:huan89215@163.com;huzhihong426@163.com
YAN Huan-huan, SHANG Yi-tong, WANG Li-hong, TIAN Xue-qin, LIAO Hai-yan, ZENG Bin, HU Zhi-hong. Heterologous Biosynthesis of Cordycepin in Aspergillus oryzae[J]. Biotechnology Bulletin, 2024, 40(6): 290-298.
| Primer name | Forward primer(5'-3') | Reverse primer(5'-3') |
|---|---|---|
| Cns1-pEX1-GFP | GAGCAGACATCACCCTCGAGATGGCCATGAACGAGAACGC | ATGGTACCTACGTACTCGAGGGCTATGCCCACCTTGGATC |
| Cns2-pEX2D-DsRed | CACAGAAGGCATTTCACGTGATGTCTTGTCCTACCAGCGC | TCCTTAAGCACGGGCACGTGTCGATGCTGCGTGCGGCTC |
| Cns3-pEX1 | GAGCAGACATCACCCTCGAGATGTCCGAGTCAACCGCCTA | ATGGTACCTACGTACTCGAGCACACGCTGATAAAGGCCGA |
| Cns3-pEX1-BFP | CCCTCGAGTACGTAGGTACCATGGTGTCTAAGGGCGAAGA | CCCTTGCTCACCATGGTACCATTAAGCTTGTGCCCCAGTT |
| Cns1-GFP/Cns2-DsRed-pEX2D | TATGACATGATTACGAATTCTCAGAGCCTAGCCAACTAGT | CTCCAGATCGCAGCGAATTCCGACGGCCAGTGCCAAGCTT |
Table 1 Primers used for vector construction
| Primer name | Forward primer(5'-3') | Reverse primer(5'-3') |
|---|---|---|
| Cns1-pEX1-GFP | GAGCAGACATCACCCTCGAGATGGCCATGAACGAGAACGC | ATGGTACCTACGTACTCGAGGGCTATGCCCACCTTGGATC |
| Cns2-pEX2D-DsRed | CACAGAAGGCATTTCACGTGATGTCTTGTCCTACCAGCGC | TCCTTAAGCACGGGCACGTGTCGATGCTGCGTGCGGCTC |
| Cns3-pEX1 | GAGCAGACATCACCCTCGAGATGTCCGAGTCAACCGCCTA | ATGGTACCTACGTACTCGAGCACACGCTGATAAAGGCCGA |
| Cns3-pEX1-BFP | CCCTCGAGTACGTAGGTACCATGGTGTCTAAGGGCGAAGA | CCCTTGCTCACCATGGTACCATTAAGCTTGTGCCCCAGTT |
| Cns1-GFP/Cns2-DsRed-pEX2D | TATGACATGATTACGAATTCTCAGAGCCTAGCCAACTAGT | CTCCAGATCGCAGCGAATTCCGACGGCCAGTGCCAAGCTT |
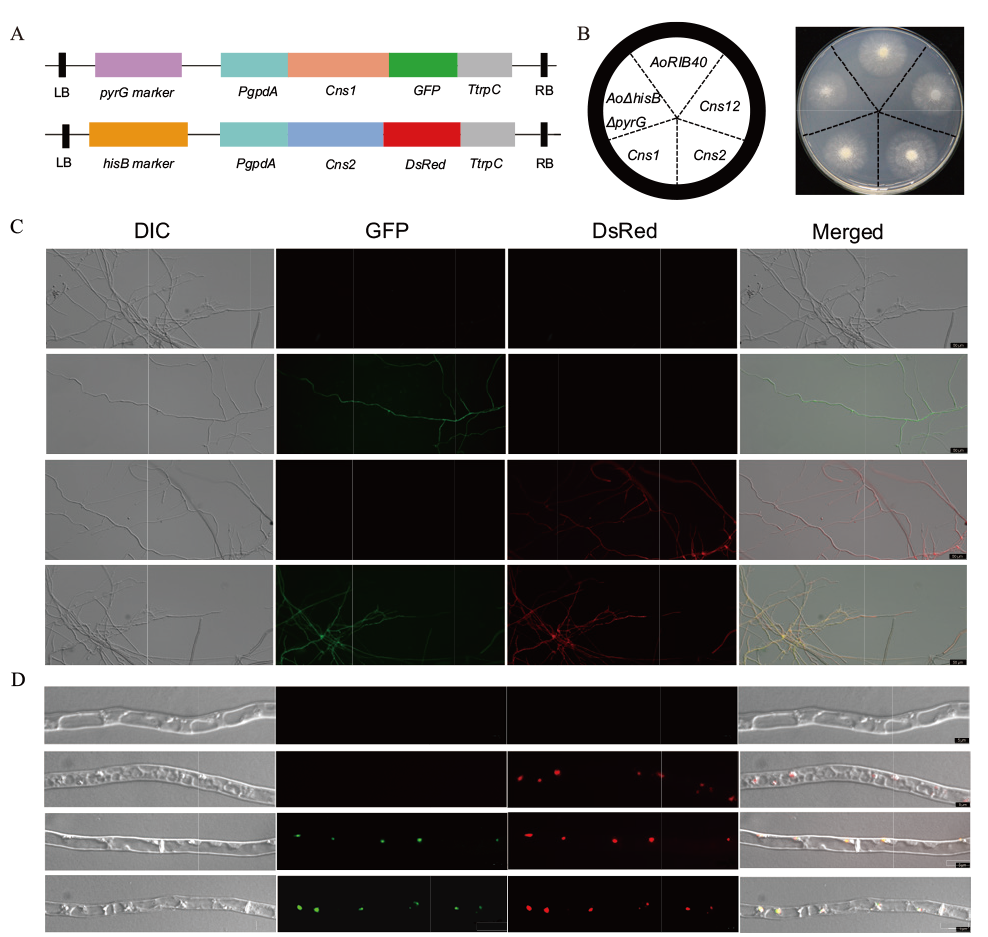
Fig. 1 Subcellular localization of CmCns1 and CmCns2 in A. oryzae A: The scheme of Cns1-pEX1-GFP and Cns2-pEX2D-DsRed overexpression vectors in A. oryzae. B: Wild-type AoRIB40, nutrient deficient strain AoΔpyrGΔHisB, transformant CmCns1/AoΔpyrGΔHisB, CmCns2/AoΔpyrGΔHisB and CmCns1 CmCns2/AoΔpyrGΔHisB were phenotyped in CD+Uri+Ura+His medium at 30℃ for 3-5 d. C: Fluorescence observation of transformants, from top to bottom: microscopic observation of mycelium of AoRIB40, transformant CmCns1/AoΔpyrGΔHisB, CmCns2/AoΔpyrGΔHisB and CmCns1 CmCns2/AoΔpyrGΔHisB in a 20-fold objective. D: Subcellular localization of CmCns1 and CmCns2 in A. oryzae, from top to bottom: microscopic observation of mycelium with AoRIB40, AoRIB40 stained with Nile red dye, transformant of CmCns1/AoΔpyrGΔHisB stained with Nile red dye, and CmCns1 CmCns2/AoΔpyrGΔHisB under 63-fold microscope. C and D: From left to right: DIC, green fluorescence of GFP, red fluorescence of DsRed, and combined images of GFP, DsRed and DIC
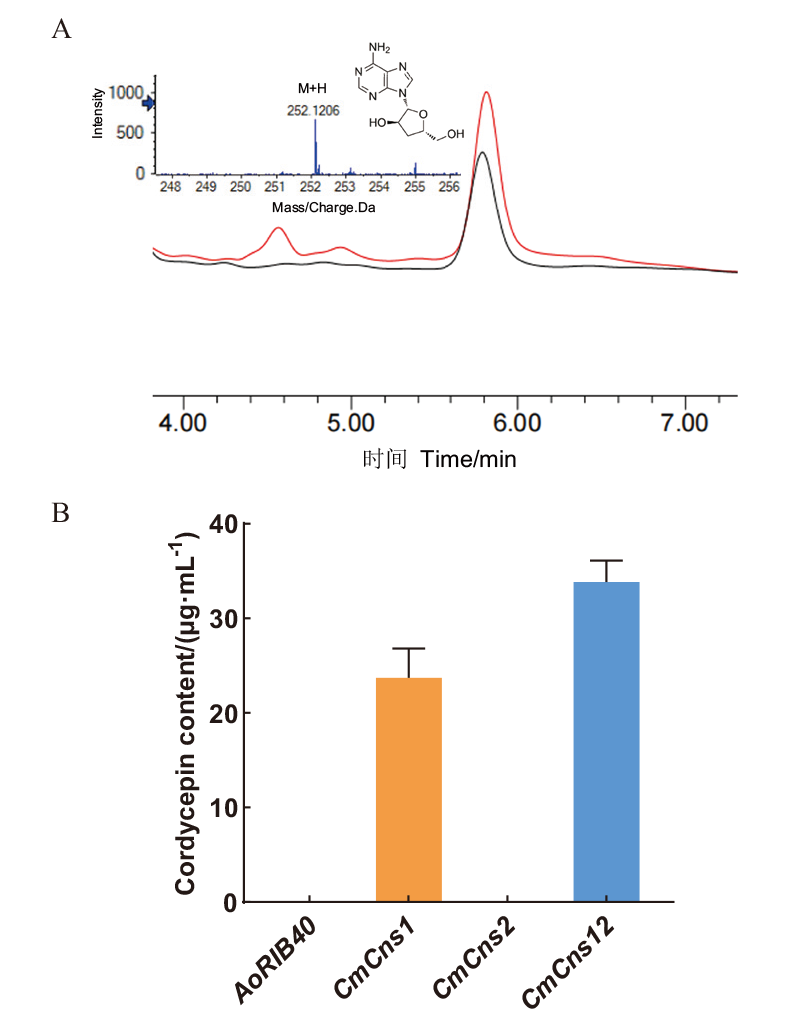
Fig. 2 Cordycepin contents of CmCns1 and CmCns2 heterologously expressed in A. oryzae A: The chromatogram peaks of cordycepin in A. oryzae transformant CmCns1/AoΔpyrGΔHisB and CmCns1 CmCns2/AoΔpyrGΔHisB by HPLC, and LC-MS/MS analysis of CmCns1 CmCns2/AoΔpyrGΔHisB. The black and red peaks respectively indicate CmCns1/AoΔpyrGΔHisB and CmCns1 CmCns2/AoΔpyrGΔHisB.B: Cordycepin contents in A. oryzae AoRIB40, heterologous expressing transformant CmCns1/AoΔpyrGΔHisB, CmCns2/AoΔpyrGΔHisB and CmCns1 CmCns2/AoΔpyrGΔHisB in liquid fermentation medium

Fig. 3 Cordycepin contents of CmCns1, CmCns2 and CmCns3 heterologously expressed in A. oryzae A: The scheme of Cns1-GFP/Cns2-DsRed-pEX2D and Cns3-pEX1-BFP vectors. B: Wild-type A. oryzae AoRIB40, background strain AoΔpyrGΔHisB, transformant CmCns1 CmCns2-pEX2D/AoΔpyrGΔHisB, CmCns3/AoΔpyrGΔHisB and CmCns1 CmCns2 CmCns3/AoΔpyrGΔHisB were phenotyped in CD+Uri+Ura+His medium at 30℃ for 3-5 d.C: From top to bottom : Microscopic observation of mycelium of transformant CmCns1CmCns2-pEX2D/AoΔpyrGΔHisB, CmCns3/AoΔpyrGΔHisB and CmCns1 CmCns2 CmCns3/AoΔpyrGΔHisB in a 40-fold objective; from left to right: DIC, green fluorescence of GFP, red fluorescence of DsRed, blue fluorescence of BFP, combined images of GFP, DsRed, BFP and DIC. D: The contents of cordycepin in A. oryzae AoRIB40 and heterologous expression transformant CmCns1/AoΔpyrGΔHisB, CmCns1 CmCns2/AoΔpyrGΔHisB and CmCns1 CmCns2 CmCns3/AoΔpyrGΔHisB liquid fermentation medium. E: The chromatogram peaks of cordycepin contents in A. oryzae transformants CmCns1/AoΔpyrGΔHisB, CmCns1 CmCns2/AoΔpyrGΔHisB and CmCns1 CmCns2 CmCns3/AoΔpyrGΔHisB by HPLC, the red, blue, and black peaks respectively correspond to CmCns1/AoΔpyrGΔHisB, CmCns1 CmCns2/AoΔpyrGΔHisB and CmCns1 CmCns2 CmCns3/AoΔpyrGΔHisB. ** indicate a significant difference compared to the control(P<0.01), the same below
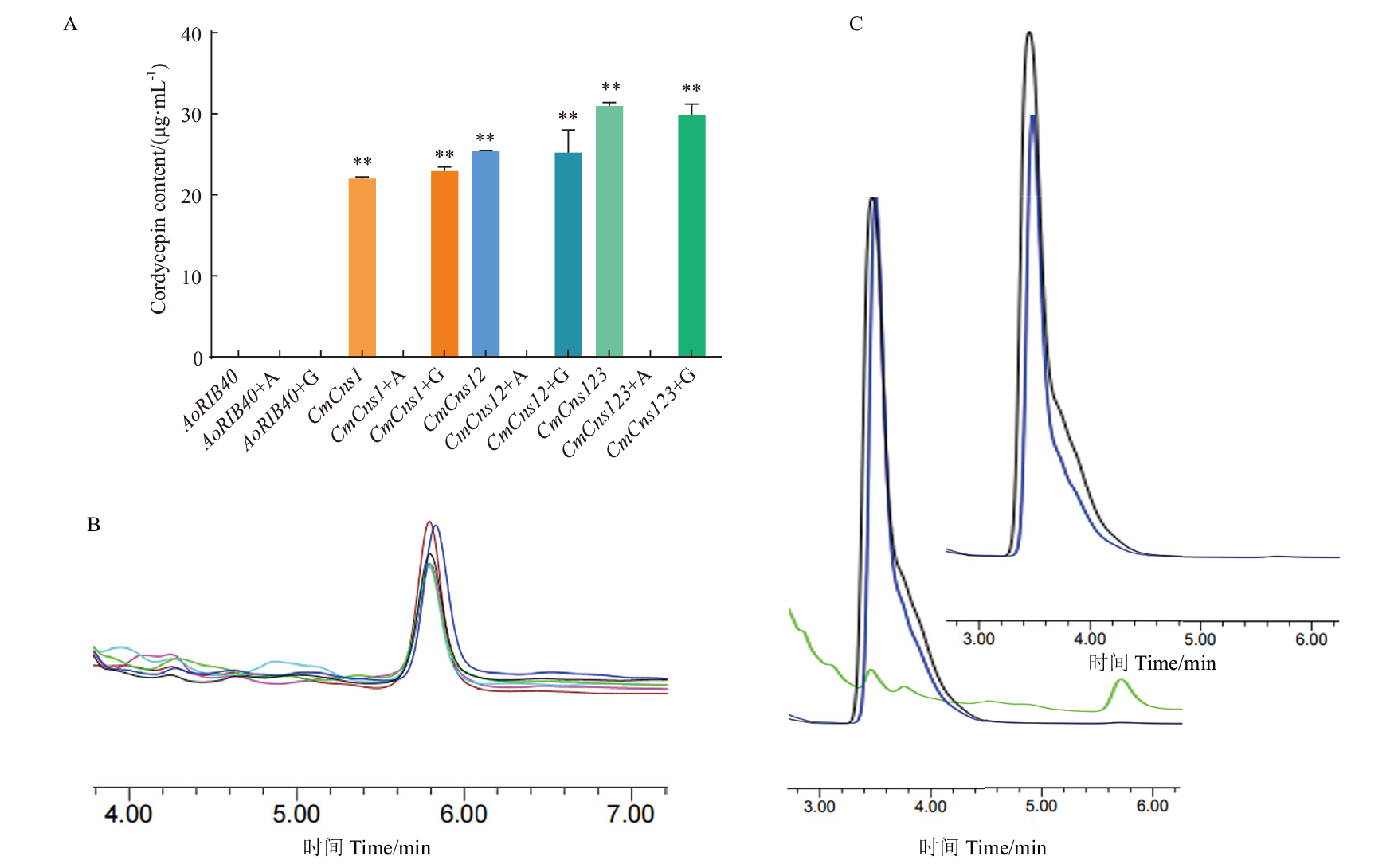
Fig. 4 Cordycepin contents of CmCns1, CmCns2 and CmCns3 heterologously expressed in A. oryzae in the liquid fermentation broth with adenine and glycine A: Cordycepin contents of A. oryzae AoRIB40, transformant CmCns1/AoΔpyrGΔHisB, CmCns1 CmCns2/AoΔpyrGΔHisB and CmCns1CmCns2 CmCns3/AoΔpyrGΔHisB in DPY medium and with glycine and adenine in the medium, respectively, AoRIB40+A indicates the addition of adenine to the medium, AoRIB40+G indicates the addition of glycine to the medium, and so on. B: Cordycepin content plots of A. oryzae transformant CmCns1/AoΔpyrGΔHisB, CmCns1 CmCns2/AoΔpyrGΔHisB, CmCns1 CmCns2 CmCns3/AoΔpyrGΔHisB and addition of glycine to the medium by HPLC, cyan, red, black, purple, green, and blue are the peaks of CmCns1/AoΔpyrGΔHisB, CmCns1 CmCns2/AoΔpyrGΔHisB, CmCns1 CmCns2 CmCns3/AoΔpyrGΔHisB and the addition of glycine, respectively. C: The chromatogram peaks of cordycepin in AoRIB40 and transformant CmCns1/AoΔpyrGΔHisB by HPLC before and after addition of adenine, the black, blue and green peaks correspond to AoRIB40, CmCns1/AoΔpyrGΔHisB after adding adenine and CmCns1/AoΔpyrGΔHisB before adding adenine, respectively
| [1] | Yang LY, Li GL, Chai Z, et al. Synthesis of cordycepin: current scenario and future perspectives[J]. Fungal Genet Biol, 2020, 143: 103431. |
| [2] |
Jin Y, Meng X, Qiu ZD, et al. Anti-tumor and anti-metastatic roles of cordycepin, one bioactive compound of Cordyceps militaris[J]. Saudi J Biol Sci, 2018, 25(5): 991-995.
doi: 10.1016/j.sjbs.2018.05.016 pmid: 30108453 |
| [3] | Taghinejad Z, Kazemi T, Fadaee M, et al. Pharmacological and therapeutic potentials of cordycepin in hematological malignancies[J]. Biochem Biophys Res Commun, 2023, 678: 135-143. |
| [4] |
Khan MA, Tania M. Cordycepin in anticancer research: molecular mechanism of therapeutic effects[J]. Curr Med Chem, 2020, 27(6): 983-996.
doi: 10.2174/0929867325666181001105749 pmid: 30277143 |
| [5] | Wang L, Yan HH, Zeng B, et al. Research progress on cordycepin synthesis and methods for enhancement of cordycepin production in Cordyceps militaris[J]. Bioengineering, 2022, 9(2): 69. |
| [6] |
Xia YL, Luo FF, Shang YF, et al. Fungal cordycepin biosynthesis is coupled with the production of the safeguard molecule pentostatin[J]. Cell Chem Biol, 2017, 24(12): 1479-1489.e4.
doi: S2451-9456(17)30327-6 pmid: 29056419 |
| [7] |
张永杰, 张姝. 蛹虫草组学研究进展[J]. 菌物学报, 2021, 40(11): 2881-2893.
doi: 10.13346/j.mycosystema.210327 |
| Zhang YJ, Zhang S. Research progress on the Cordyceps militaris omics[J]. Mycosystema, 2021, 40(11): 2881-2893. | |
| [8] | 霍春红, 李鸿宇, 李倩, 等. 产虫草素酿酒酵母工程菌株的构建与发酵优化[J]. 生物工程学报, 2021, 37(9): 3334-3347. |
| Huo CH, Li HY, Li Q, et al. Construction and optimization of cordycepin-producing Saccharomyces cerevisiae[J]. Chin J Biotechnol, 2021, 37(9): 3334-3347. | |
| [9] | Song ZQ, Lin WB, Duan XY, et al. Increased cordycepin production in Yarrowia lipolytica using combinatorial metabolic engineering strategies[J]. ACS Synth Biol, 2023, 12(3): 780-787. |
| [10] | Yu JY, Sun M, Wang XY, et al. Poly-pathways metabolomics for high-yielding cordycepin of Cordyceps militaris[J]. Biomed Chromatogr, 2023, 37(2): e5551. |
| [11] | Masuda M, Urabe E, Honda H, et al. Enhanced production of cordycepin by surface culture using the medicinal mushroom Cordyceps militaris[J]. Enzyme Microb Technol, 2007, 40(5): 1199-1205. |
| [12] | Sari N, Suparmin A, Kato T, et al. Improved cordycepin production in a liquid surface culture of Cordyceps militaris isolated from wild strain[J]. Biotechnol Bioprocess Eng, 2016, 21(5): 595-600. |
| [13] | Machida M, Yamada O, Gomi K. Genomics of Aspergillus oryzae: learning from the history of Koji mold and exploration of its future[J]. DNA Res, 2008, 15(4): 173-183. |
| [14] | Wang L, Hu TT, Jiang ZQ, et al. Efficient production of a novel alkaline cold-active phospholipase C from Aspergillus oryzae by molecular chaperon co-expression for crude oil degumming[J]. Food Chem, 2021, 350: 129212. |
| [15] | Machida M, Asai K, Sano M, et al. Genome sequencing and analysis of Aspergillus oryzae[J]. Nature, 2005, 438(7071): 1157-1161. |
| [16] | Fujii R, Minami A, Tsukagoshi T, et al. Total biosynthesis of diterpene aphidicolin, a specific inhibitor of DNA polymerase α: heterologous expression of four biosynthetic genes in Aspergillus oryzae[J]. Biosci Biotechnol Biochem, 2011, 75(9): 1813-1817. |
| [17] | Ban A, Tanaka M, Fujii R, et al. Subcellular localization of aphidicolin biosynthetic enzymes heterologously expressed in Aspergillus oryzae[J]. Biosci Biotechnol Biochem, 2018, 82(1): 139-147. |
| [18] | Ugai T, Minami A, Fujii R, et al. Heterologous expression of highly reducing polyketide synthase involved in betaenone biosynthesis[J]. Chem Commun, 2015, 51(10): 1878-1881. |
| [19] | Tagami K, Minami A, Fujii R, et al. Rapid reconstitution of biosynthetic machinery for fungal metabolites in Aspergillus oryzae: total biosynthesis of aflatrem[J]. Chembiochem, 2014, 15(14): 2076-2080. |
| [20] | Feng J, Hauser M, Cox RJ, et al. Engineering Aspergillus oryzae for the heterologous expression of a bacterial modular polyketide synthase[J]. J Fungi, 2021, 7(12): 1085. |
| [21] | Thai HD, Nguyen BPT, Nguyen VM, et al. Development of a new Agrobacterium-mediated transformation system based on a dual auxotrophic approach in the filamentous fungus Aspergillus oryzae[J]. World J Microbiol Biotechnol, 2021, 37(6): 92. |
| [22] | Sun YL, Niu YL, Huang H, et al. Mevalonate diphosphate decarboxylase MVD/Erg19 is required for ergosterol biosynthesis, growth, sporulation and stress tolerance in Aspergillus oryzae[J]. Front Microbiol, 2019, 10: 1074. |
| [23] | Jin Q, Li GH, Qin KH, et al. The expression pattern, subcellular localization and function of three sterol 14α-demethylases in Aspergillus oryzae[J]. Front Genet, 2023, 14: 1009746. |
| [24] | Hu ZH, Huang H, Sun YL, et al. Effects on gene transcription profile and fatty acid composition by genetic modification of mevalonate diphosphate decarboxylase MVD/Erg19 in Aspergillus oryzae[J]. Microorganisms, 2019, 7(9): 342. |
| [25] | 闫欢欢, 尚怡彤, 王丽红, 等. 蛹虫草磷酸甲羟戊酸激酶和焦磷酸甲羟戊酸脱羧酶基因的功能分析[J]. 微生物学报, 2024, 64(2): 461-472. |
| Yan HH, Shang YT, Wang LH, et al. Functions of genes encoding phosphomevalonate kinase and mevalonate diphosphate decarboxylase in Cordyceps militaris[J]. Acta Microbiologica Sinica, 2024, 64(2): 461-472. | |
| [26] | Nguyen KT, Ho QN, Pham TH, et al. The construction and use of versatile binary vectors carrying pyrG auxotrophic marker and fluorescent reporter genes for Agrobacterium-mediated transformation of Aspergillus oryzae[J]. World J Microbiol Biotechnol, 2016, 32(12): 204. |
| [27] | 王立, 黄慧, 刘新平, 等. 农杆菌介导的蛹虫草营养缺陷型菌株和遗传转化体系的构建[J]. 微生物学通报, 2022, 49(8): 3373-3386. |
| Wang L, Huang H, Liu XP, et al. Construction of Agrobacterium-mediated auxotrophic strain and genetic transformation system of Cordyceps militaris[J]. Microbiol China, 2022, 49(8): 3373-3386. | |
| [28] | Chen M, Luo JH, Jiang WM, et al. Cordycepin: a review of strategies to improve the bioavailability and efficacy[J]. Phytother Res, 2023, 37(9): 3839-3858. |
| [29] | Turk A, Abdelhamid MAA, Yeon SW, et al. Cordyceps mushroom with increased cordycepin content by the cultivation on edible insects[J]. Front Microbiol, 2022, 13: 1017576. |
| [30] | Turk A, Lee S, Yeon SW, et al. Adenosine deaminase inhibitory activity of medicinal plants: boost the production of cordycepin in Cordyceps militaris[J]. Antioxidants, 2023, 12(6): 1260. |
| [31] | Duan XY, Yang H, Wang C, et al. Microbial synthesis of cordycepin, current systems and future perspectives[J]. Trends Food Sci Technol, 2023, 132: 162-170. |
| [32] | Borde M, Singh SK. Enhanced production of cordycepin under solid-state fermentation of Cordyceps militaris by using combinations of grains/substrates[J]. Braz J Microbiol, 2023, 54(4): 2765-2772. |
| [33] |
Alberti F, Foster GD, Bailey AM. Natural products from filamentous fungi and production by heterologous expression[J]. Appl Microbiol Biotechnol, 2017, 101(2): 493-500.
doi: 10.1007/s00253-016-8034-2 pmid: 27966047 |
| [1] | LI Bo-jing, ZHENG La-mei, WU Wu-yun, GAO Fei, ZHOU Yi-jun. Evolution, Expression, and Functional Analysis of the HSP20 Gene Family from Simmondisa chinensis [J]. Biotechnology Bulletin, 2024, 40(6): 190-202. |
| [2] | WU Ze-hang, YANG Zhong-yi, YAN Yi-cheng, JIA Yong-hong, WU Yue-yan, XIE Xiao-hong. Cloning and Functional Analysis of Flavonoid 3'-hydroxylase(F3'H)Gene in Rhododendron hybridum Hort [J]. Biotechnology Bulletin, 2024, 40(6): 251-259. |
| [3] | LI Meng-ran, YE Wei, LI Sai-ni, ZHANG Wei-yang, LI Jian-jun, ZHANG Wei-min. Expression of Lithocarols Biosynthesis Gene litI and Functional Analysis of Its Promoter [J]. Biotechnology Bulletin, 2024, 40(6): 310-318. |
| [4] | PAN Ping-ping, XU Zhi-hao, ZHANG Yi-wen, LI Qing, WANG Zhong-hua. Prokaryotic Expression, Subcellular Localization and Expression Analysis of PcCHS Gene from Polygonatum cyrtonema Hua [J]. Biotechnology Bulletin, 2024, 40(5): 280-289. |
| [5] | ZHANG Zhen, LI Qing, XU Jing, CHEN Kai-yuan, ZHANG Chun-zhi, ZHU Guang-tao. Construction and Application of Potato Mitochondrial Targeted Expression Vector [J]. Biotechnology Bulletin, 2024, 40(5): 66-73. |
| [6] | YIN Liang, WANG Dai-wei, LIU Yue-ying, LIU Hai-yan, LUO Guang-hong. Cloning and Expression of Protease SpP1 Gene and Characterization of Enzymatic Properties [J]. Biotechnology Bulletin, 2024, 40(4): 278-286. |
| [7] | YANG Yan, HU Yang, LIU Ni-ru, YIN Lu, YANG Rui, WANG Peng-fei, MU Xiao-peng, ZHANG Shuai, CHENG Chun-zhen, ZHANG Jian-cheng. Cloning and Functional Analysis of MbbZIP43 Gene in ‘Hongmantang’ Red-flesh Apple [J]. Biotechnology Bulletin, 2024, 40(2): 146-159. |
| [8] | LI Hao, WU Guo-qiang, WEI Ming, HAN Yue-xin. Genome-wide Identification of the BvBADH Gene Family in Sugar Beet(Beta vulgaris)and Their Expression Analysis Under High Salt Stress [J]. Biotechnology Bulletin, 2024, 40(2): 233-244. |
| [9] | ZHENG Fei, YANG Jun-zhao, NIU Yu-feng, LI Rui-lin, ZHAO Guo-zhu. Characterization and Functional Analysis of Lytic Polysaccharide Monooxygenase TtLPMO9I from Thermothelomyces thermophilus [J]. Biotechnology Bulletin, 2024, 40(2): 289-299. |
| [10] | ZHU Yi, LIU Tang-jing, GONG Guo-yi, ZHANG Jie, WANG Jin-fang, ZHANG Hai-ying. Cloning and Expression Analysis of ClPP2C3 in Citrullus lanatus [J]. Biotechnology Bulletin, 2024, 40(1): 243-249. |
| [11] | XIE Hong, ZHOU Li-ying, LI Shu-wen, WANG Meng-di, AI Ye, CHAO Yue-hui. Structural and Functional Analysis of MtCIM Gene in Medicago truncatula [J]. Biotechnology Bulletin, 2024, 40(1): 262-269. |
| [12] | LYU Qiu-yu, SUN Pei-yuan, RAN Bin, WANG Jia-rui, CHEN Qing-fu, LI Hong-you. Cloning, Subcellular Localization and Expression Analysis of the Transcription Factor Gene FtbHLH3 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 194-203. |
| [13] | LI Bo, LIU He-xia, CHEN Yu-ling, ZHOU Xing-wen, ZHU Yu-lin. Cloning, Subcellular Localization and Expression Analysis of CnbHLH79 Transcription Factor from Camellia nitidissima [J]. Biotechnology Bulletin, 2023, 39(8): 241-250. |
| [14] | WANG Shuai, FENG Yu-mei, BAI Miao, DU Wei-jun, YUE Ai-qin. Functional Analysis of Soybean Gene GmHMGR Responding to Exogenous Hormones and Abiotic Stresses [J]. Biotechnology Bulletin, 2023, 39(7): 131-142. |
| [15] | MA Yu-qian, SUN Dong-hui, YUE Hao-feng, XIN Jia-yu, LIU Ning, CAO Zhi-yan. Identification, Heterologous Expression and Functional Analysis of a GH61 Family Glycoside Hydrolase from Setosphaeria turcica with the Assisting Function in Degrading Cellulose [J]. Biotechnology Bulletin, 2023, 39(4): 124-135. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||