








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (6): 310-318.doi: 10.13560/j.cnki.biotech.bull.1985.2023-1225
Previous Articles Next Articles
LI Meng-ran1,2( ), YE Wei2, LI Sai-ni2, ZHANG Wei-yang2, LI Jian-jun1(
), YE Wei2, LI Sai-ni2, ZHANG Wei-yang2, LI Jian-jun1( ), ZHANG Wei-min2(
), ZHANG Wei-min2( )
)
Received:2023-12-29
Online:2024-06-26
Published:2024-06-24
Contact:
LI Jian-jun, ZHANG Wei-min
E-mail:1871288385@qq.com;lijianjun1672@163.com;wmzhang@gdim.cn
LI Meng-ran, YE Wei, LI Sai-ni, ZHANG Wei-yang, LI Jian-jun, ZHANG Wei-min. Expression of Lithocarols Biosynthesis Gene litI and Functional Analysis of Its Promoter[J]. Biotechnology Bulletin, 2024, 40(6): 310-318.
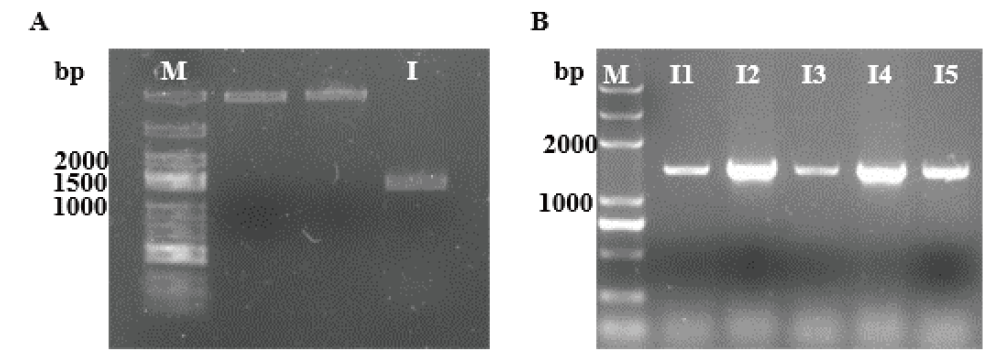
Fig. 2 PCR verification of gene litI A: Cloning of gene litI, M: DL5000 DNA marker; I: gene litI. B: PCR verification of bacterial liquid, M: DL5000 DNA marke; I1-5: positive colonies of pET-30a-litI
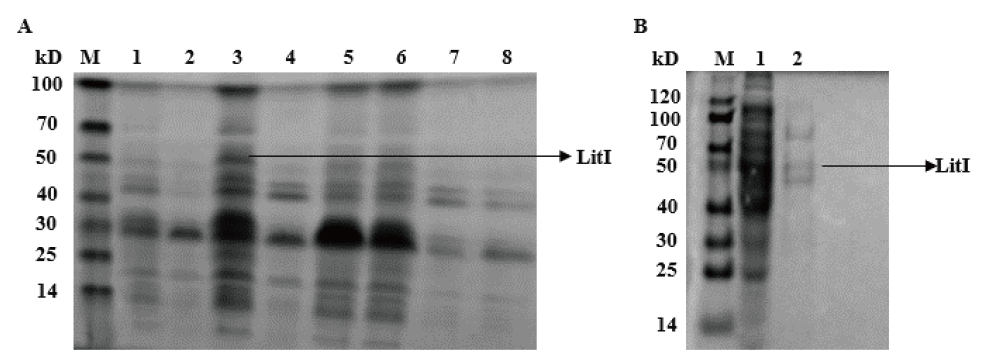
Fig. 3 Preliminary purification of protein LitI A: Optimization of protein LitI expression conditions. M: Blue Plus II protein marker(14-100 kD); 1, 3, 5 and 6: supernatant for ①, ③, ② and ④; 2, 4, 7 and 8: sediment for ①, ③, ② and ④. B: Preliminary purification of protein LitI, M: protein marker; 1: binding buffer; 2: 10% imidazole eluent
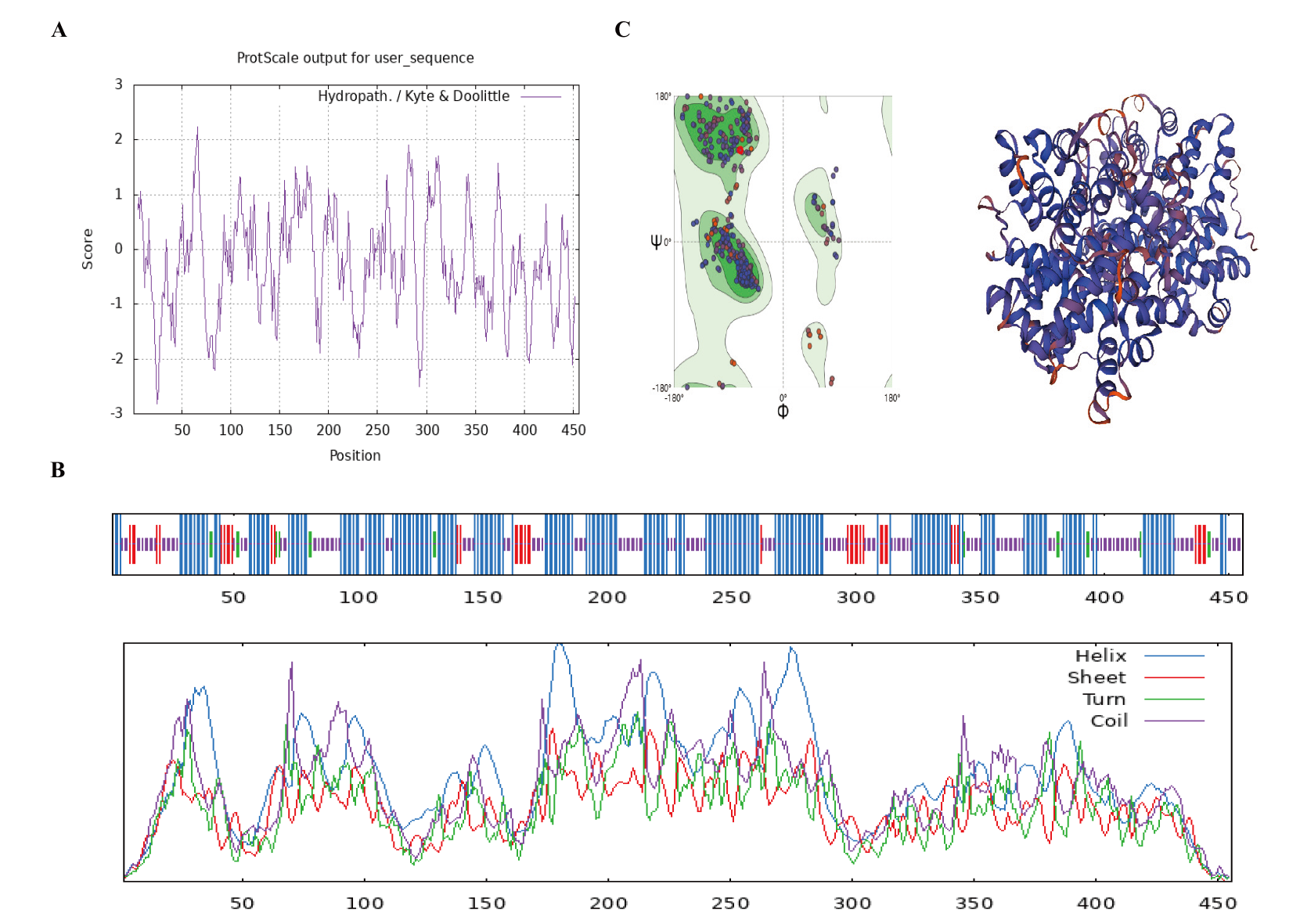
Fig.4 Bioinformatics analysis of LitI protein A: Hydrophilicity/hydrophobicity analysis of protein LitI. B: Secondary structure analysis of protein LitI. C: Homology modeling for tertiary structure prediction of protein LitI
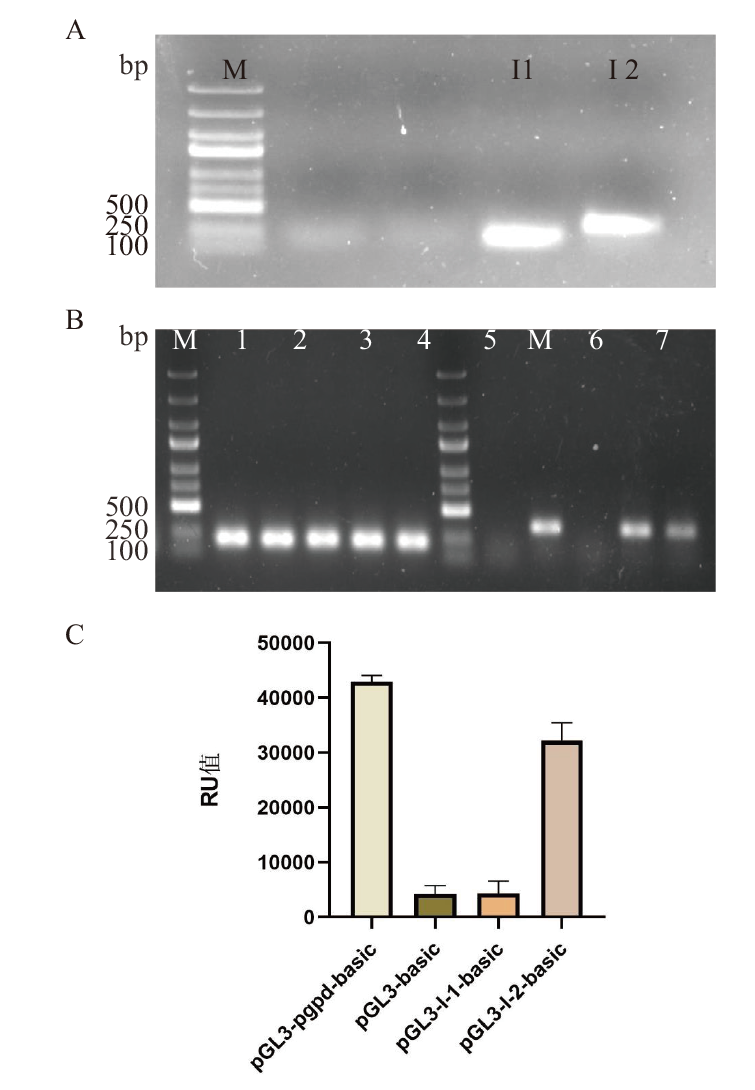
Fig. 5 Construction of pGL3-basic-promoter recombinant vector and analysis of its transcriptional activity A: Cloning of the litI promoter fragments; M: DL5000 DNA marker; I-1, I-2: promoter fragments amplified by PCR. B: PCR validation of colonies with pGL3-basic-litI promoter; M: DL5000 DNA marker; 1-5: PCR validation of colonies containing I-1 promoter; 6-10: PCR validation of colonies containing I-2 promoter. C: Luciferase RU values of the pGL3-basic-unloaded/pgpd/litI promoter vector in Trans5α cells
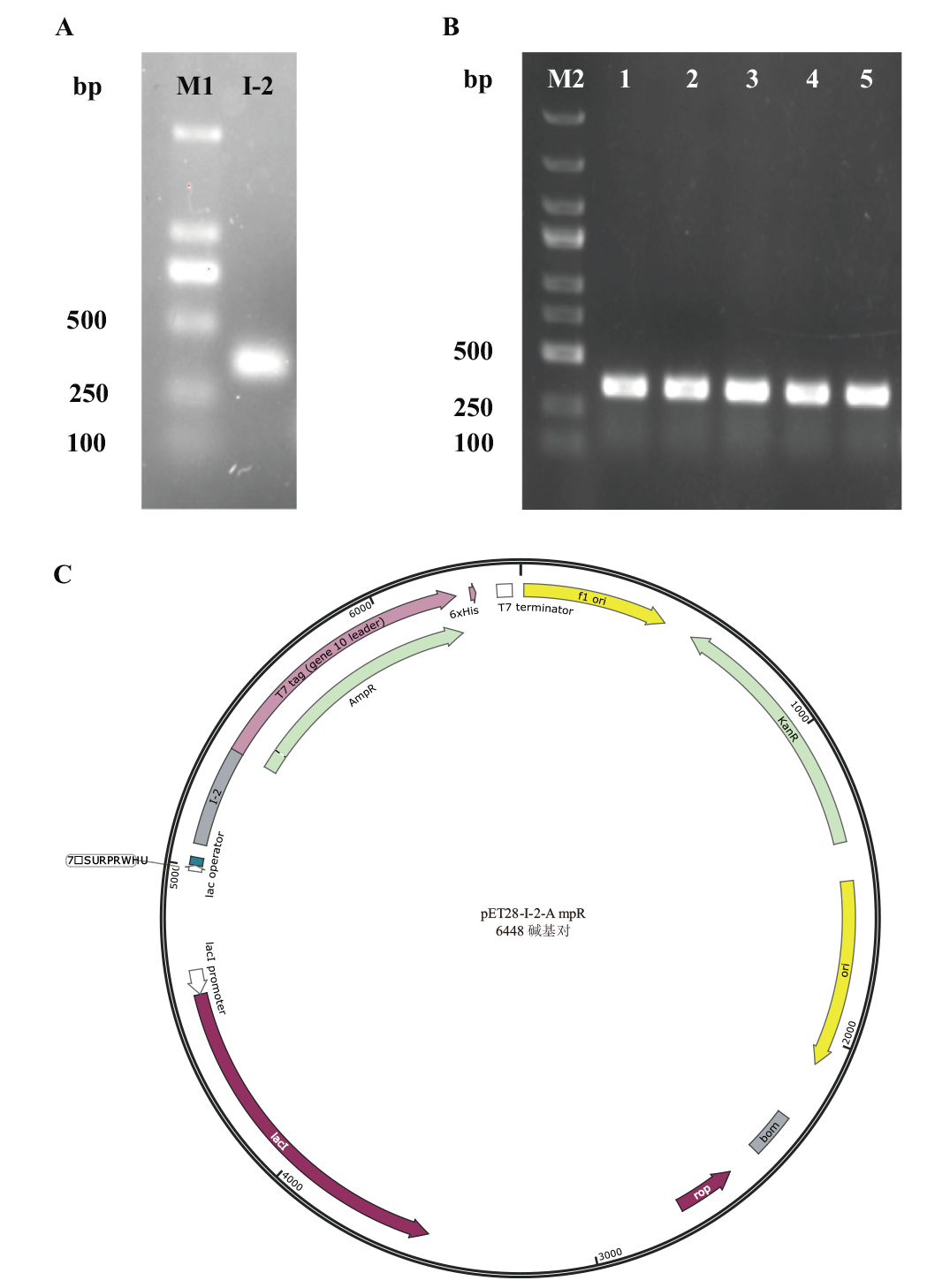
Fig.6 PCR verification of Trans5α colony and promoter vector map A: litI promoter fragment amplified with -pET28a-Amp homology arm primers; M1: Trans2K DNA marker. B: Colony PCR of Trans5α-pET28a-I-2; M2: DL5000 DNA marker; C: vector map of Trans5α-pET28a-I-2-AmpR

Fig.7 Growth of E. coli containing promoter fragments on ampicillin-resistant plates A: The plates of Trans5α-pEASY-T1-AmpR, Trans5α-pET28a-ACP-AmpR, and Trans5α-pET28a-I-2-AmpR, P: Trans5α-pEASY-T1-AmpR(Positive control); N: Trans5α-pET28a-ACP-AmpR(Negative control); I: Trans5α-pET28a-I-2-AmpR. B: The OD values of Trans5α-pET28a-ACP-AmpR, Trans5α-pEASY-T1-AmpR and Trans5α-pET28a-I-2-AmpR in LB medium at 0, 25, and 40 μg/mL AMP, respectively
| 启动子片段 Promoter fragment | 启动子序列 Promoter sequence(5'-3') |
|---|---|
| I-2 | gcttgaatgcagcaagcaggcaactcgggcatcggagttgaccaatttggga caggcggaggtacatgtacccatgtagccatgcatacggccacctcccaag ataaatcacagctgatgatcagaccatattgctgagcatgttcgacaaacg ttttctgcgcagttagccgctatAAAAAtgttaatagaaggtaattcgatc gaaaccgtttgcaaataatattcagtgccacaagtcggtcttcaccggctc catacgattattcgcagatactgagacgacatactatctgttcattctcacctgaacatg |
Table 1 Sequence of litI promoter fragment I-2
| 启动子片段 Promoter fragment | 启动子序列 Promoter sequence(5'-3') |
|---|---|
| I-2 | gcttgaatgcagcaagcaggcaactcgggcatcggagttgaccaatttggga caggcggaggtacatgtacccatgtagccatgcatacggccacctcccaag ataaatcacagctgatgatcagaccatattgctgagcatgttcgacaaacg ttttctgcgcagttagccgctatAAAAAtgttaatagaaggtaattcgatc gaaaccgtttgcaaataatattcagtgccacaagtcggtcttcaccggctc catacgattattcgcagatactgagacgacatactatctgttcattctcacctgaacatg |
| [1] | Zhang CW, Ondeyka JG, Herath KB, et al. Tenellones A and B from a Diaporthe sp.: two highly substituted benzophenone inhibitors of parasite cGMP-dependent protein kinase activity[J]. J Nat Prod, 2005, 68(4): 611-613. |
| [2] | Cui H, Lin Y, Luo MC, et al. Diaporisoindoles A-C: three isoprenylisoindole alkaloid derivatives from the mangrove endophytic fungus Diaporthe sp. SYSU-HQ3[J]. Org Lett, 2017, 19(20): 5621-5624. |
| [3] | Cui H, Liu YN, Li J, et al. Diaporindenes A-D: four unusual 2, 3-dihydro-1 H-indene analogues with anti-inflammatory activities from the mangrove endophytic fungus Diaporthe sp. SYSU-HQ3[J]. J Org Chem, 2018, 83(19): 11804-11813. |
| [4] |
Gonzalez-Andrade M, Rivera-Chavez J, Sosa-Peinado A, et al. Development of the fluorescent biosensor hCalmodulin(hCaM)L39C-monobromobimane(mBBr)/V91C-mBBr, a novel tool for discovering new calmodulin inhibitors and detecting calcium[J]. J Med Chem, 2011, 54(11): 3875-3884.
doi: 10.1021/jm200167g pmid: 21495717 |
| [5] | Holker JSE, Lapper RD, Simpson TJ. The biosynthesis of fungal metabolites. Part IV. Tajixanthone: 13C nuclear magnetic resonance spectrum and feedings with[1-13C]- and[2-13C]-acetate[J]. J Chem Soc, Perkin Trans 1, 1974: 2135. |
| [6] | Ishida M, Hamasaki T, Hatsuda Y. The structure of two new metabolites, emerin and emericellin, from Aspergillus nidulans[J]. Agricultural and Biological Chemistry, 1975, 39(11): 2181-2184. |
| [7] | Ishida M, Hamasaki T, Hatsuda Y. Biosynthesis of shamixanthone[J]. Agricultural and Biological Chemistry, 1978, 42(2): 465-466. |
| [8] | Simpson TJ, Stenzel DJ. Biosynthesis of aflatoxins. Incorporation of[1, 2-13C2]acetate,[2H3]acetate, and[1-13C, 2H3]acetate into sterigmatocystin in Aspergillus versicolor[J]. J Chem Soc, Chem Commun, 1982(15): 890-892. |
| [9] | Xu JL, Liu ZM, Chen YC, et al. Lithocarols A-F, six tenellone derivatives from the deep-sea derived fungus Phomopsis lithocarpus FS508[J]. Bioorg Chem, 2019, 87: 728-735. |
| [10] |
Simpson TJ. Genetic and biosynthetic studies of the fungal prenylated xanthone shamixanthone and related metabolites in Aspergillus spp. revisited[J]. Chembiochem, 2012, 13(11): 1680-1688.
doi: 10.1002/cbic.201200014 pmid: 22730213 |
| [11] |
郑禾, 周玉珂, 林贤福, 等. Baeyer-Villiger单加氧酶的蛋白质改造及其催化氧化反应研究新进展[J]. 有机化学, 2019, 39(4): 903-915.
doi: 10.6023/cjoc201810023 |
| Zheng H, Zhou YK, Lin XF, et al. Recent developments in protein engineering and catalytic oxidations of baeyer-villiger monooxygenase[J]. Chin J Org Chem, 2019, 39(4): 903-915. | |
| [12] | Ceccoli RD, Bianchi DA, Fink MJ, et al. Cloning and characterization of the Type I Baeyer-Villiger monooxygenase from Leptospira biflexa[J]. AMB Express, 2017, 7(1): 87. |
| [13] |
Riebel A, Dudek HM, de Gonzalo G, et al. Expanding the set of rhodococcal Baeyer-Villiger monooxygenases by high-throughput cloning, expression and substrate screening[J]. Appl Microbiol Biotechnol, 2012, 95(6): 1479-1489.
doi: 10.1007/s00253-011-3823-0 pmid: 22218769 |
| [14] |
刘小琴, 杨岩, 吴喆瑜, 等. 多点突变提高α-L-鼠李糖苷酶热稳定性[J]. 食品与发酵工业, 2019, 45(6): 23-29.
doi: 10.13995/j.cnki.11-1802/ts.018703 |
| Liu XQ, Yang Y, Wu ZY, et al. Enhanced thermostability of α-L-rhamnosidase by multiple-site mutation[J]. Food Ferment Ind, 2019, 45(6): 23-29. | |
| [15] | 朱士臣, 冯媛, 刘书来, 等. 鱼糜凝胶热稳定性的增强技术研究进展[J]. 中国食品学报, 2022, 22(7): 384-396. |
| Zhu SC, Feng Y, Liu SL, et al. Recent advances of technologies to enhance thermal stability of surimi gel products[J]. J Chin Inst Food Sci Technol, 2022, 22(7): 384-396. | |
| [16] | 杜昊澜, 骆树娟, Tosin Victor Adegoke, 等. 基于热不稳定区段分析和二硫键预测的玉米赤霉烯酮脱毒酶ZLHY-6热稳定性改造[J]. 食品安全质量检测学报, 2022, 13(22): 7150-7157. |
| Du HL, Luo SJ, Adegoke TV, et al. Thermal stability modification of Zearalenone detoxification enzyme ZLHY-6 based on thermolabile segment analysis and disulfide bond prediction[J]. J Food Saf Qual, 2022, 13(22): 7150-7157. | |
| [17] |
Liu L, Liu J, Qiu RX, et al. Improving heterologous gene expression in Aspergillus niger by introducing multiple copies of protein-binding sequence containing CCAAT to the promoter[J]. Lett Appl Microbiol, 2003, 36(6): 358-361.
pmid: 12753242 |
| [18] | Ashrafi M, Farsi M, Mirshamsi A, et al. Isolation and sequence analysis of gpdII promoter of the white button mushroom(Agaricus bisporus)from strains Holland737 and IM008[J]. International Journal of Horticultural Science and Technology, 2015, 2(1):33-41. |
| [1] | CHANG Xue-rui, WANG Tian-tian, WANG Jing. Identification and Analysis of E2 Gene Family in Pepper(Capsicum annuum L.) [J]. Biotechnology Bulletin, 2024, 40(6): 238-250. |
| [2] | LIU Rong, TIAN Min-yu, LI Guang-ze, TAN Cheng-fang, RUAN Ying, LIU Chun-lin. Identification and Induced-expression Analysis of REVEILLE Family in Brassica napus L. [J]. Biotechnology Bulletin, 2024, 40(6): 161-171. |
| [3] | HU Yong-bo, LEI Yu-tian, YANG Yong-sen, CHEN Xin, LIN Huang-fang, LIN Bi-ying, LIU Shuang, BI Ge, SHEN Bao-ying. Genome-wide Identification and Expression Pattern Analysis of the Bcl-2-related Anti-apoptotic Family in Cucumis sativus L. and Cucurbita moschata Duch. [J]. Biotechnology Bulletin, 2024, 40(6): 219-237. |
| [4] | YAN Huan-huan, SHANG Yi-tong, WANG Li-hong, TIAN Xue-qin, LIAO Hai-yan, ZENG Bin, HU Zhi-hong. Heterologous Biosynthesis of Cordycepin in Aspergillus oryzae [J]. Biotechnology Bulletin, 2024, 40(6): 290-298. |
| [5] | WANG Jian, YANG Sha, SUN Qing-wen, CHEN Hong-yu, YANG Tao, HUANG Yuan. Genome-wide Identification and Expression Analysis of bHLH Transcription Factor Family in Dendrobium nobile [J]. Biotechnology Bulletin, 2024, 40(6): 203-218. |
| [6] | WANG Zhou, YU Jie, WANG Jin-hua, WANG Yong-ze, ZHAO Xiao. Anaerobic Expression of Lactate Dehydrogenase to Improve the D-lactic Acid Optical Purity Procluced by Escherichia coli [J]. Biotechnology Bulletin, 2024, 40(5): 290-299. |
| [7] | HOU Ya-qiong, LANG Hong-shan, WEN Meng-meng, GU Yi-yun, ZHU Run-jie, TANG Xiao-li. Identification and Expression Analysis of AcHSP20 Gene Family in Kiwifruit [J]. Biotechnology Bulletin, 2024, 40(5): 167-178. |
| [8] | LI Jia-xin, LI Hong-yan, LIU Li-e, ZHANG Tian, ZHOU Wu. Identification and Expression Analysis of the NRAMP Family in Seabuckthorn Under Lead Stress [J]. Biotechnology Bulletin, 2024, 40(5): 191-202. |
| [9] | XIAO Ya-ru, JIA Ting-ting, LUO Dan, WU Zhe, LI Li-xia. Cloning and Expression Analysis of CsERF025L Transcription Factor in Cucumber [J]. Biotechnology Bulletin, 2024, 40(4): 159-166. |
| [10] | ZHONG Yun, LIN Chun, LIU Zheng-jie, DONG Chen-wen-hua, MAO Zi-chao, LI Xing-yu. Cloning and Prokaryotic Expression Analysis of Asparagus Saponin Synthesis Related Glycosyltransferase Genes [J]. Biotechnology Bulletin, 2024, 40(4): 255-263. |
| [11] | YIN Liang, WANG Dai-wei, LIU Yue-ying, LIU Hai-yan, LUO Guang-hong. Cloning and Expression of Protease SpP1 Gene and Characterization of Enzymatic Properties [J]. Biotechnology Bulletin, 2024, 40(4): 278-286. |
| [12] | ZHANG Qing-lan, ZHANG Ya-ran, JU Zhi-hua, WANG Xiu-ge, XIAO Yao, WANG Jin-peng, WEI Xiao-chao, GAO Ya-ping, BAI Fu-heng, WANG Hong-cheng. Identification and Transcriptional Regulation Analysis of Core Promoter in Bovine TARDBP Gene [J]. Biotechnology Bulletin, 2024, 40(4): 306-318. |
| [13] | CHEN Xiao-song, LIU Chao-jie, ZHENG Jia, QIAO Zong-wei, LUO Hui-bo, ZOU Wei. Analyzing the Growth and Caproic Acid Metabolism Mechanism of Rummeliibacillus suwonensis 3B-1 by Tandem Mass Tag-based Quantitative Proteomics [J]. Biotechnology Bulletin, 2024, 40(3): 135-145. |
| [14] | YANG Wei-jie, YANG Zhou-lin, ZHU Hao-dong, WEI Yu, LIU Jun, LIU Xun. Study on the Properties and Functions of LchAD Protein, a Key Module of Lichenysin Synthase [J]. Biotechnology Bulletin, 2024, 40(3): 322-332. |
| [15] | GONG Li-li, YU Hua, YANG Jie, CHEN Tian-chi, ZHAO Shuang-ying, WU Yue-yan. Identification and Analysis of Grape(Vitis vinifera L.)CYP707A Gene Family and Functional Verification to Fruit Ripening [J]. Biotechnology Bulletin, 2024, 40(2): 160-171. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||