








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (8): 83-94.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0156
Previous Articles Next Articles
LIN Tong1( ), YUAN Cheng1, DONG Chen-wen-hua1,2, ZENG Meng-qiong1, YANG Yan1, MAO Zi-chao1,2, LIN Chun1,2(
), YUAN Cheng1, DONG Chen-wen-hua1,2, ZENG Meng-qiong1, YANG Yan1, MAO Zi-chao1,2, LIN Chun1,2( )
)
Received:2024-02-15
Online:2024-08-26
Published:2024-09-05
Contact:
LIN Chun
E-mail:941985193@qq.com;linchun@ynau.edu.cn
LIN Tong, YUAN Cheng, DONG Chen-wen-hua, ZENG Meng-qiong, YANG Yan, MAO Zi-chao, LIN Chun. Screening and Functional Analysis of Gene CqSTK Associated with Gametophyte Development of Quinoa[J]. Biotechnology Bulletin, 2024, 40(8): 83-94.
| 引物名称Primer name | 引物序列Primer sequence(5'-3') | 用途Usage |
|---|---|---|
| CqSTK F | ATGGGGAGAGGGAAGATAGAG | 基因克隆Gene cloning |
| CqSTK R | TTACCTTAGGTGAAATAGCTTCTTC | 基因克隆Gene cloning |
| Cqactin F | GTCCACAGAAAGTGCTTCTAAG | 内参基因Acting |
| Cqactin R | AACAACTCCTCACCTTCTCATG | 内参基因Acting |
| CqSTKqpcr F1 | ATAGAGAACACGACGAATCGT | RT-qPCR |
| CqSTKqpcr R1 | AACCAAATACAGATGATGCAA | RT-qPCR |
Table 1 Primer sequence
| 引物名称Primer name | 引物序列Primer sequence(5'-3') | 用途Usage |
|---|---|---|
| CqSTK F | ATGGGGAGAGGGAAGATAGAG | 基因克隆Gene cloning |
| CqSTK R | TTACCTTAGGTGAAATAGCTTCTTC | 基因克隆Gene cloning |
| Cqactin F | GTCCACAGAAAGTGCTTCTAAG | 内参基因Acting |
| Cqactin R | AACAACTCCTCACCTTCTCATG | 内参基因Acting |
| CqSTKqpcr F1 | ATAGAGAACACGACGAATCGT | RT-qPCR |
| CqSTKqpcr R1 | AACCAAATACAGATGATGCAA | RT-qPCR |
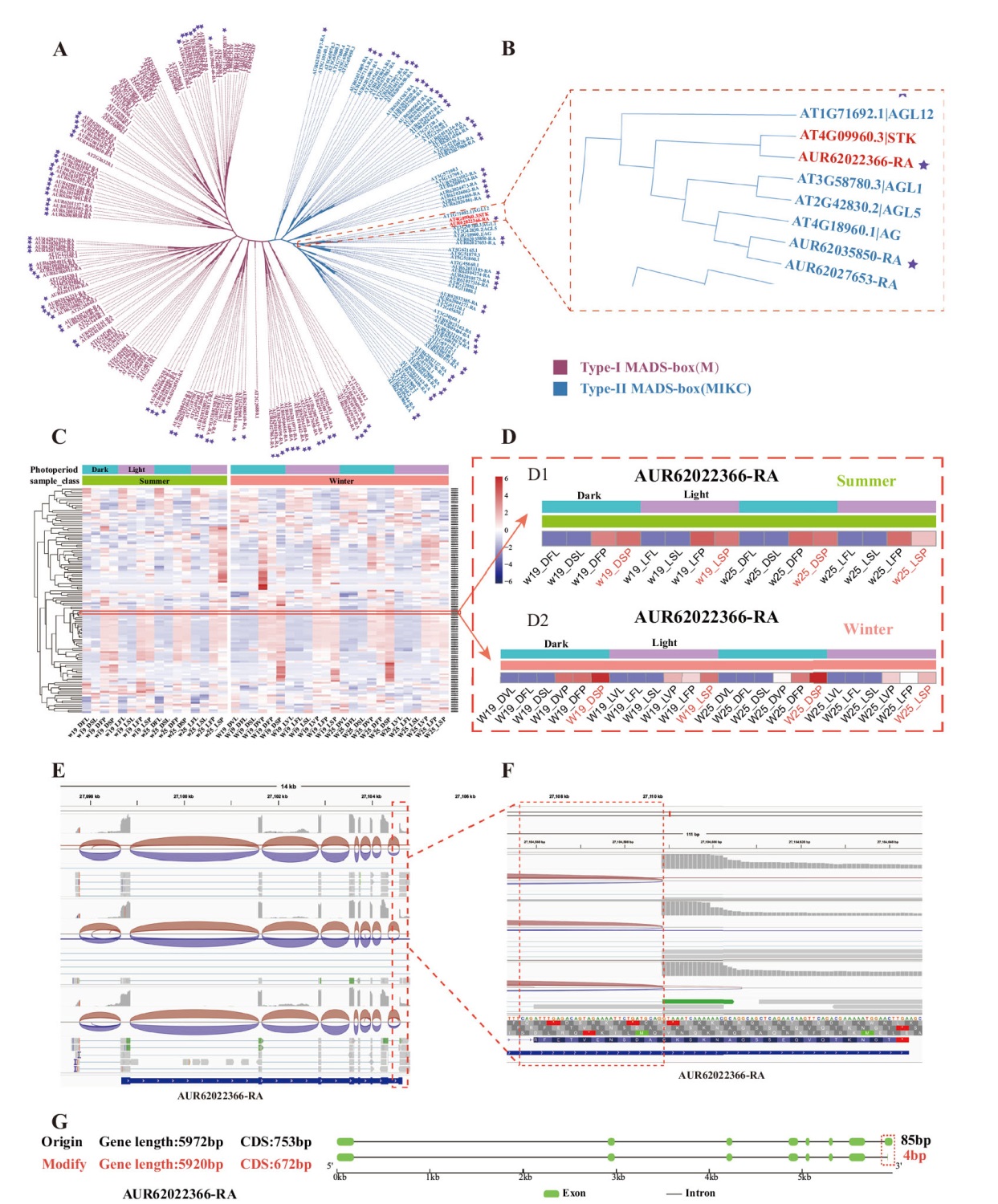
Fig. 1 Identification and sequence optimization of the CqSTK in quinoa A: Phylogenetic tree of quinoa and Arabidopsis MADS-box gene family members(purple pentagrams indicate quinoa MADS-box members). B: Differential gene AUR62022366-RA clusters with Arabidopsis STK gene. C, D: Heatmaps of MADS-box gene family expressions. E, F: IGV visualization plots. G: Gene structure diagrams. DVL refers to short-day treatment vegetative leaves, LVL to long-day treatment vegetative leaves, DVP to short-day treatment vegetative inflorescence, LVP to long-day treatment vegetative inflorescence, DFL to short-day treatment flowering phase leaves, LFL to long-day treatment flowering phase leaves. DFP refers to short-day treatment flowering phase inflorescence, LFP to long-day treatment flowering phase inflorescence. DSL refers to short-day treatment ripening phase leaves, LSL to long-day treatment ripening phase leaves; DSP to short-day treatment ripening phase inflorescence, LSP to long-day treatment ripening phase inflorescence
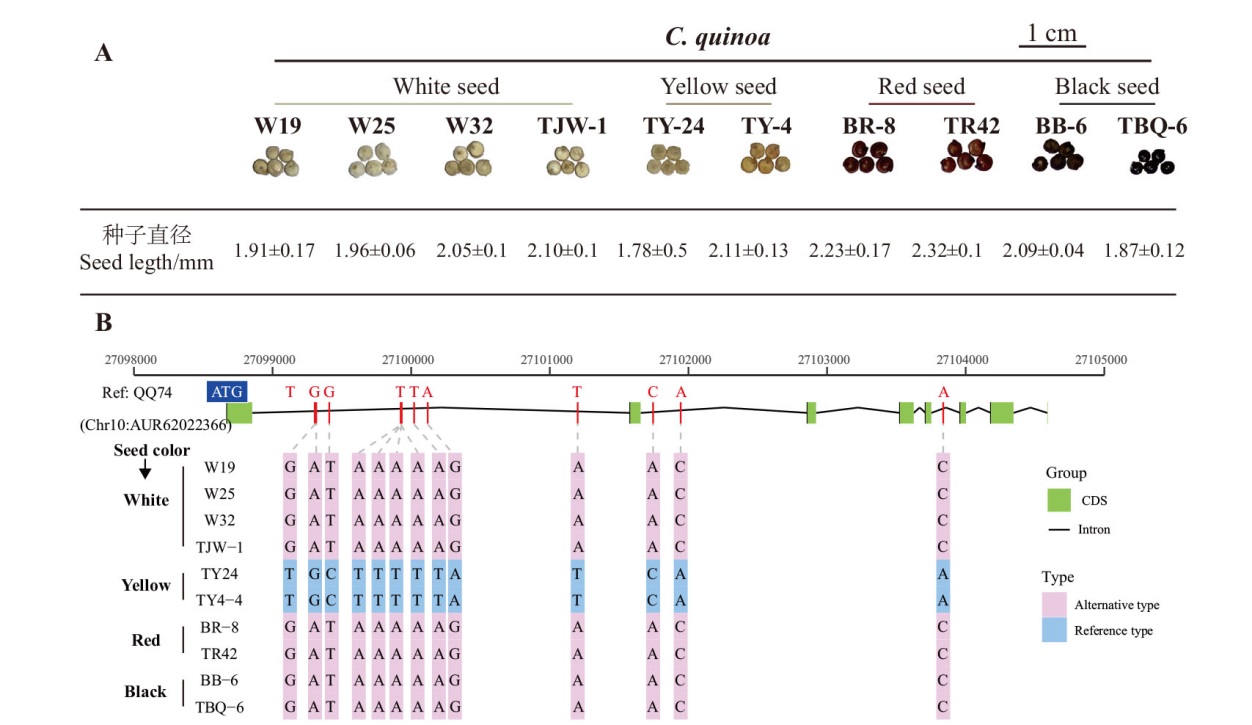
Fig. 2 Phenotypic characteristics of 10 quinoa germplasms and the information on CqSTK allele genes A: Seed phenotype statistics of the 10 quinoa varieties, categorized into four groups: white, yellow, red, and black. B: Sequence information of CqSTK in the 10 varieties with QQ74 as the reference genome. The green boxes(exons)and black lines(introns)form the gene structure of CqSTK. Red font indicates the bases at specific loci of the reference genome QQ74, pink background denotes inconsistencies with the reference genome, and blue background signifies consistency with the reference genome
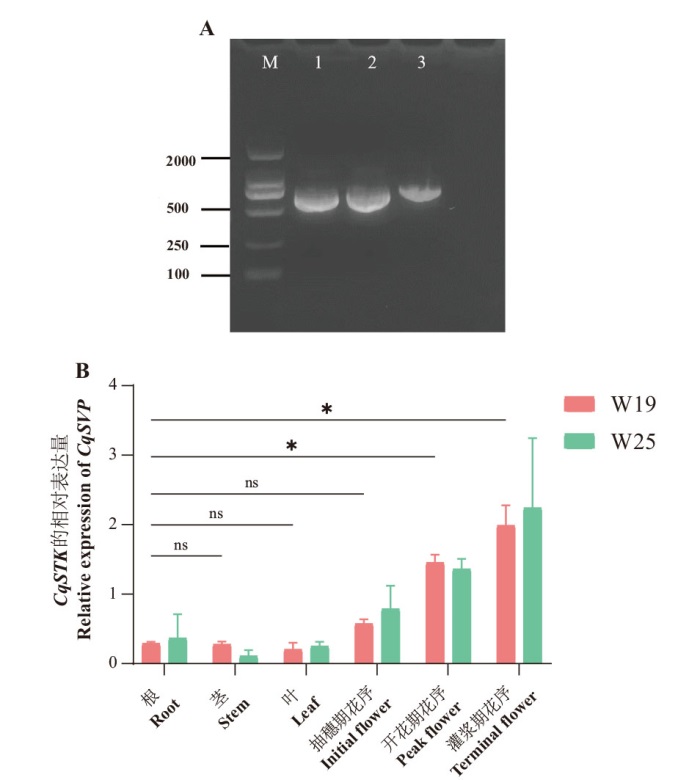
Fig. 4 Cloning and spatiotemporal expression of CqSTK A: Full-length CDS cloning of CqSTK using cDNA as a template. B: Expression levels of CqSTK in different varieties and tissues, * indicates significant difference(P < 0.05), ns indicates no significant difference
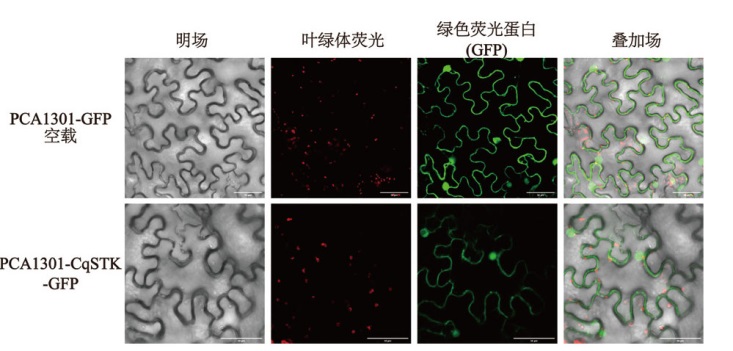
Fig. 5 Subcellular localization of CqSTK PCA1301-eGFP enables transient expression of green fluorescent protein(GFP)in tobacco cells. PCA1301-CqSTK-GFP is a fusion expression vector containing both CqSTK and GFP, allowing for the simultaneous expression of CqSTK and GFP. The localization of CqSTK expression can be indirectly inferred by detecting the position of green fluorescence. Chloroplast fluorescence refers to the red fluorescence emitted by chloroplasts at specific wavelengths. The overlay image consists of three fields: bright field, chloroplast fluorescence, and green fluorescent protein
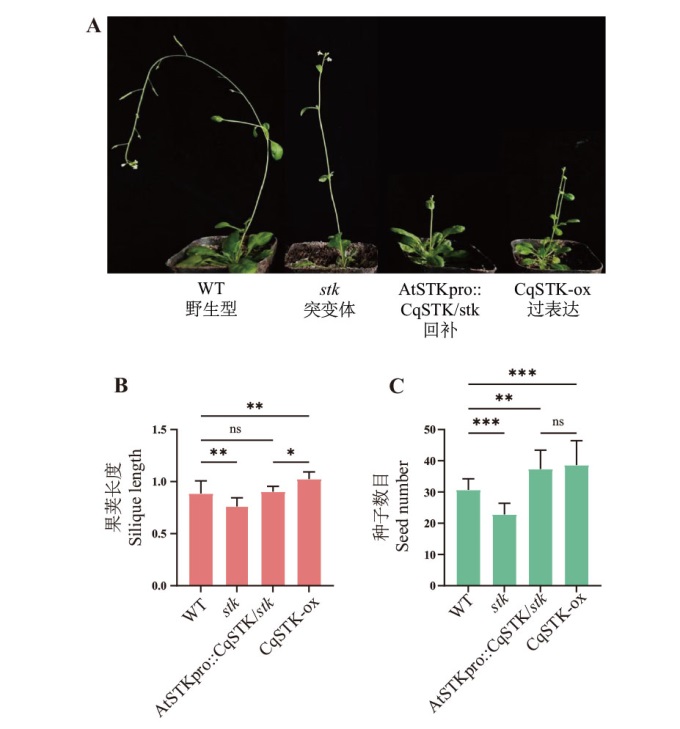
Fig. 6 Phenotypic variations in Arabidopsis and transgenic materials A: Morphology of wild-type Arabidopsis, stk mutant, overexpressing T3 generation, and stk mutant complementation T3 generation plants. B: Silique length. C: Number of seeds per silique. *P<0.05, **P<0.01, ***P<0.001, and ns indicates no significant difference
| 材料 Material | 首次开花时间 First flowering time/d | 首次开花莲座叶片 Number of first flowering lotus leaves/pieces |
|---|---|---|
| 拟南芥野生型 Wild type | 45 | 20 |
| 拟南芥stk突变体 stk mutant | 49 | 16 |
| 回补CqSTK/stk Complementation CqSTK in stk mutant | 72 | 28 |
| 过表达CqSTK Overexpress CqSTK in wild type | 61 | 25 |
Table 2 First flowering time and rosette leaf number of wild-type and transgenic Arabidopsis
| 材料 Material | 首次开花时间 First flowering time/d | 首次开花莲座叶片 Number of first flowering lotus leaves/pieces |
|---|---|---|
| 拟南芥野生型 Wild type | 45 | 20 |
| 拟南芥stk突变体 stk mutant | 49 | 16 |
| 回补CqSTK/stk Complementation CqSTK in stk mutant | 72 | 28 |
| 过表达CqSTK Overexpress CqSTK in wild type | 61 | 25 |
| [1] | Zurita-Silva A, Fuentes F, Zamora P, et al. Breeding quinoa(Chenopodium quinoa Willd.): potential and perspectives[J]. Mol Breed, 2014, 34(1): 13-30. |
| [2] |
Hariadi Y, Marandon K, Tian Y, et al. Ionic and osmotic relations in quinoa(Chenopodium quinoa Willd.)plants grown at various salinity levels[J]. J Exp Bot, 2011, 62(1): 185-193.
doi: 10.1093/jxb/erq257 pmid: 20732880 |
| [3] | Ruales J, Nair BM. Content of fat, vitamins and minerals in quinoa(Chenopodium quinoa, Willd)seeds[J]. Food Chem, 1993, 48(2): 131-136. |
| [4] | Adolf VI, Shabala S, Andersen MN, et al. Varietal differences of quinoa's tolerance to saline conditions[J]. Plant Soil, 2012, 357(1): 117-129. |
| [5] | Jacobsen SE, Mujica A, Jensen CR. The resistance of quinoa(Chenopodium quinoaWilld.)to adverse abiotic factors[J]. Food Rev Int, 2003, 19(1/2): 99-109. |
| [6] | Alandia G, Rodriguez JP, Jacobsen SE, et al. Global expansion of quinoa and challenges for the Andean region[J]. Glob Food Secur, 2020, 26: 100429. |
| [7] | Losa A, Colombo M, Brambilla V, et al. Genetic interaction between AINTEGUMENTA(ANT)and the ovule identity genes SEEDSTICK(STK), SHATTERPROOF1(SHP1)and SHATTERPROOF2(SHP2)[J]. Sex Plant Reprod, 2010, 23(2): 115-121. |
| [8] |
Dreni L, Kater MM. MADS reloaded: evolution of the AGAMOUS subfamily genes[J]. New Phytol, 2014, 201(3): 717-732.
doi: 10.1111/nph.12555 pmid: 24164649 |
| [9] | Mizzotti C, Mendes MA, Caporali E, et al. The MADS box genes SEEDSTICK and ARABIDOPSIS Bsister play a maternal role in fertilization and seed development[J]. Plant J, 2012, 70(3): 409-420. |
| [10] | Pinyopich A, Ditta GS, Savidge B, et al. Assessing the redundancy of MADS-box genes during carpel and ovule development[J]. Nature, 2003, 424(6944): 85-88. |
| [11] | Di Marzo M, Herrera-Ubaldo H, Caporali E, et al. SEEDSTICK controls Arabidopsis fruit size by regulating cytokinin levels and FRUITFULL[J]. Cell Rep, 2020, 30(8): 2846-2857.e3. |
| [12] | Paolo D, Orozco-Arroyo G, Rotasperti L, et al. Genetic interaction of SEEDSTICK, GORDITA and AUXIN RESPONSE FACTOR 2 during seed development[J]. Genes, 2021, 12(8): 1189. |
| [13] |
Balanzà V, Roig-Villanova I, Di Marzo M, et al. Seed abscission and fruit dehiscence required for seed dispersal rely on similar genetic networks[J]. Development, 2016, 143(18): 3372-3381.
doi: 10.1242/dev.135202 pmid: 27510967 |
| [14] |
Fischer A, Baum N, Saedler H, et al. Chromosomal mapping of the MADS-box multigene family in Zea mays reveals dispersed distribution of allelic genes as well as transposed copies[J]. Nucleic Acids Res, 1995, 23(11): 1901-1911.
doi: 10.1093/nar/23.11.1901 pmid: 7596816 |
| [15] |
Schmidt RJ, Veit B, Mandel MA, et al. Identification and molecular characterization of ZAG1, the maize homolog of the Arabidopsis floral homeotic gene AGAMOUS[J]. Plant Cell, 1993, 5(7): 729-737.
doi: 10.1105/tpc.5.7.729 pmid: 8103379 |
| [16] |
Ambrose BA, Lerner DR, Ciceri P, et al. Molecular and genetic analyses of the silky1 gene reveal conservation in floral organ specification between eudicots and monocots[J]. Mol Cell, 2000, 5(3): 569-579.
doi: 10.1016/s1097-2765(00)80450-5 pmid: 10882141 |
| [17] | Dreni L, Pilatone A, Yun DP, et al. Functional analysis of all AGAMOUS subfamily members in rice reveals their roles in reproductive organ identity determination and meristem determinacy[J]. Plant Cell, 2011, 23(8): 2850-2863. |
| [18] | Ocarez N, Mejía N. Suppression of the D-class MADS-box AGL11 gene triggers seedlessness in fleshy fruits[J]. Plant Cell Rep, 2016, 35(1): 239-254. |
| [19] | Wang QJ, Dan NZ, Zhang XN, et al. Identification, characterization and functional analysis of C-class genes associated with double flower trait in carnation(Dianthus caryphyllus L.)[J]. Plants, 2020, 9(1): 87. |
| [20] | Yellina AL, Orashakova S, Lange S, et al. Floral homeotic C function genes repress specific B function genes in the carpel whorl of the basal eudicot California poppy(Eschscholzia californica)[J]. EvoDevo, 2010, 1: 13. |
| [21] | Serwatowska J, Roque E, Gómez-Mena C, et al. Two euAGAMOUS genes control C-function in Medicago truncatula[J]. PLoS One, 2014, 9(8): e103770. |
| [22] | 袁加红, 刘正杰, 吴慧琳, 等. 111份藜麦种质资源农艺性状分析[J]. 云南农业大学学报: 自然科学, 2020, 35(4): 572-580, 650. |
| Yuan JH, Liu ZJ, Wu HL, et al. The analysis of the main agronomic traits of 111 quinoa germplasm resources[J]. J Yunnan Agric Univ Nat Sci, 2020, 35(4): 572-580, 650. | |
| [23] |
Gramzow L, Theissen G. A hitchhiker's guide to the MADS world of plants[J]. Genome Biol, 2010, 11(6): 214.
doi: 10.1186/gb-2010-11-6-214 pmid: 20587009 |
| [24] |
Smaczniak C, Immink RGH, Angenent GC, et al. Developmental and evolutionary diversity of plant MADS-domain factors: insights from recent studies[J]. Development, 2012, 139(17): 3081-3098.
doi: 10.1242/dev.074674 pmid: 22872082 |
| [25] |
Alvarez J, Smyth DR. CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS[J]. Development, 1999, 126(11): 2377-2386.
doi: 10.1242/dev.126.11.2377 pmid: 10225997 |
| [26] | Coen ES, Meyerowitz EM. The war of the whorls: genetic interactions controlling flower development[J]. Nature, 1991, 353(6339): 31-37. |
| [27] | Pelaz S, Ditta GS, Baumann E, et al. B and C floral organ identity functions require SEPALLATA MADS-box genes[J]. Nature, 2000, 405(6783): 200-203. |
| [28] |
Schneitz K. The molecular and genetic control of ovule development[J]. Curr Opin Plant Biol, 1999, 2(1): 13-17.
doi: 10.1016/s1369-5266(99)80003-x pmid: 10047571 |
| [29] |
Theissen G, Kim JT, Saedler H. Classification and phylogeny of the MADS-box multigene family suggest defined roles of MADS-box gene subfamilies in the morphological evolution of eukaryotes[J]. J Mol Evol, 1996, 43(5): 484-516.
pmid: 8875863 |
| [30] | Nguyen LT, Schmidt HA, von Haeseler A, et al. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies[J]. Mol Biol Evol, 2015, 32(1): 268-274. |
| [31] | Robinson JT, Thorvaldsdottir H, Turner D, et al. Igv. js: an embeddable JavaScript implementation of the Integrative Genomics Viewer(IGV)[J]. Bioinformatics, 2023, 39(1): btac830. |
| [32] | 张东亮. 藜麦MADS-box基因家族的鉴定与分析及发根农杆菌介导的根转化研究[D]. 烟台: 烟台大学, 2021. |
| Zhang DL. Identification and analysis of quinoa MADS-box gene family and study on Agrobacterium-mediated root transformation[D]. Yantai: Yantai University, 2021. | |
| [33] |
Malabarba J, Buffon V, Mariath JEA, et al. The MADS-box gene Agamous-like 11 is essential for seed morphogenesis in grapevine[J]. J Exp Bot, 2017, 68(7): 1493-1506.
doi: 10.1093/jxb/erx025 pmid: 28369525 |
| [34] |
Colombo L, Franken J, Van der Krol AR, et al. Downregulation of ovule-specific MADS box genes from petunia results in maternally controlled defects in seed development[J]. Plant Cell, 1997, 9(5): 703-715.
pmid: 9165748 |
| [35] |
Costantini L, Battilana J, Lamaj F, et al. Ber1ry and phenology-related traits in grapevine(Vitis vinifera L.): from quantitative trait loci to underlying genes[J]. BMC Plant Biol, 2008, 8: 38.
doi: 10.1186/1471-2229-8-38 pmid: 18419811 |
| [36] | Ocarez N, Mejía N. Suppression of the D-class MADS-box AGL11 gene triggers seedlessness in fleshy fruits[J]. Plant Cell Rep, 2016, 35(1): 239-254. |
| [37] |
Kobayashi Y, Kaya H, Goto K, et al. A pair of related genes with antagonistic roles in mediating flowering signals[J]. Science, 1999, 286(5446): 1960-1962.
doi: 10.1126/science.286.5446.1960 pmid: 10583960 |
| [38] |
Zicola J, Liu LY, Tänzler P, et al. Targeted DNA methylation represses two enhancers of FLOWERING LOCUS T in Arabidopsis thaliana[J]. Nat Plants, 2019, 5(3): 300-307.
doi: 10.1038/s41477-019-0375-2 pmid: 30833712 |
| [39] | Suárez-López P, Wheatley K, Robson F, et al. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis[J]. Nature, 2001, 410(6832): 1116-1120. |
| [40] |
Samach A, Onouchi H, Gold SE, et al. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis[J]. Science, 2000, 288(5471): 1613-1616.
doi: 10.1126/science.288.5471.1613 pmid: 10834834 |
| [41] | Gregis V, Andrés F, Sessa A, et al. Identification of pathways directly regulated by short vegetative phase during vegetative and reproductive development in Arabidopsis[J]. Genome Biol, 2013, 14(6): R56. |
| [42] | Simonini S, Roig-Villanova I, Gregis V, et al. Basic pentacysteine proteins mediate MADS domain complex binding to the DNA for tissue-specific expression of target genes in Arabidopsis[J]. Plant Cell, 2012, 24(10): 4163-4172. |
| [43] | Matias-Hernandez L, Battaglia R, Galbiati F, et al. VERDANDI is a direct target of the MADS domain ovule identity complex and affects embryo sac differentiation in Arabidopsis[J]. Plant Cell, 2010, 22(6): 1702-1715. |
| [44] | Paolo D, Rotasperti L, Schnittger A, et al. The Arabidopsis MADS-domain transcription factor SEEDSTICK controls seed size via direct activation of E2Fa[J]. Plants, 2021, 10(2): 192. |
| [45] |
He SC, Min YC, Liu ZJ, et al. Antagonistic MADS-box transcription factors SEEDSTICK and SEPALLATA3 form a transcriptional regulatory network that regulates seed oil accumulation[J]. J Integr Plant Biol, 2024, 66(1): 121-142.
doi: 10.1111/jipb.13606 |
| [1] | XING Li-nan, ZHANG Yan-fang, GE Ming-ran, ZHAO Ling-min, CHEN Yan, HUO Xiu-wen. Analysis of DoWRKY40 Gene Expression Characteristics and Screening of Interacting Proteins in Yam [J]. Biotechnology Bulletin, 2024, 40(8): 118-128. |
| [2] | SUN Hui-qiong, ZHANG Chun-lai, WANG Xi-liang, XU Hong-shen, DOU Miao-miao, YANG Bo-hui, CHAI Wen-ting, ZHAO Shan-shan, JIANG Xiao-dong. Identification, Expression and DNA Variation Analysis of FLS Gene Family in Chenopodium quinoa [J]. Biotechnology Bulletin, 2024, 40(7): 172-182. |
| [3] | LI Bo-jing, ZHENG La-mei, WU Wu-yun, GAO Fei, ZHOU Yi-jun. Evolution, Expression, and Functional Analysis of the HSP20 Gene Family from Simmondisa chinensis [J]. Biotechnology Bulletin, 2024, 40(6): 190-202. |
| [4] | WU Ze-hang, YANG Zhong-yi, YAN Yi-cheng, JIA Yong-hong, WU Yue-yan, XIE Xiao-hong. Cloning and Functional Analysis of Flavonoid 3'-hydroxylase(F3'H)Gene in Rhododendron hybridum Hort [J]. Biotechnology Bulletin, 2024, 40(6): 251-259. |
| [5] | YAN Huan-huan, SHANG Yi-tong, WANG Li-hong, TIAN Xue-qin, LIAO Hai-yan, ZENG Bin, HU Zhi-hong. Heterologous Biosynthesis of Cordycepin in Aspergillus oryzae [J]. Biotechnology Bulletin, 2024, 40(6): 290-298. |
| [6] | PAN Ping-ping, XU Zhi-hao, ZHANG Yi-wen, LI Qing, WANG Zhong-hua. Prokaryotic Expression, Subcellular Localization and Expression Analysis of PcCHS Gene from Polygonatum cyrtonema Hua [J]. Biotechnology Bulletin, 2024, 40(5): 280-289. |
| [7] | ZHANG Zhen, LI Qing, XU Jing, CHEN Kai-yuan, ZHANG Chun-zhi, ZHU Guang-tao. Construction and Application of Potato Mitochondrial Targeted Expression Vector [J]. Biotechnology Bulletin, 2024, 40(5): 66-73. |
| [8] | YANG Yan, HU Yang, LIU Ni-ru, YIN Lu, YANG Rui, WANG Peng-fei, MU Xiao-peng, ZHANG Shuai, CHENG Chun-zhen, ZHANG Jian-cheng. Cloning and Functional Analysis of MbbZIP43 Gene in ‘Hongmantang’ Red-flesh Apple [J]. Biotechnology Bulletin, 2024, 40(2): 146-159. |
| [9] | ZHU Yi, LIU Tang-jing, GONG Guo-yi, ZHANG Jie, WANG Jin-fang, ZHANG Hai-ying. Cloning and Expression Analysis of ClPP2C3 in Citrullus lanatus [J]. Biotechnology Bulletin, 2024, 40(1): 243-249. |
| [10] | XIE Hong, ZHOU Li-ying, LI Shu-wen, WANG Meng-di, AI Ye, CHAO Yue-hui. Structural and Functional Analysis of MtCIM Gene in Medicago truncatula [J]. Biotechnology Bulletin, 2024, 40(1): 262-269. |
| [11] | LYU Qiu-yu, SUN Pei-yuan, RAN Bin, WANG Jia-rui, CHEN Qing-fu, LI Hong-you. Cloning, Subcellular Localization and Expression Analysis of the Transcription Factor Gene FtbHLH3 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 194-203. |
| [12] | LI Bo, LIU He-xia, CHEN Yu-ling, ZHOU Xing-wen, ZHU Yu-lin. Cloning, Subcellular Localization and Expression Analysis of CnbHLH79 Transcription Factor from Camellia nitidissima [J]. Biotechnology Bulletin, 2023, 39(8): 241-250. |
| [13] | TENG Meng-xin, XU Ya, HE Jing, WANG Qi, QIAO Fei, LI Jing-yang, LI Xin-guo. Cloning and Prokaryotic Expression Analysis of MaMC6 in Banana [J]. Biotechnology Bulletin, 2023, 39(12): 179-186. |
| [14] | SHANG Yi-tong, YAN Huan-huan, WANG Li-hong, TIAN Xue-qin, XUE Ping-hong, LUO Tao, HU Zhi-hong. Study on the Function of Phosphomevalonate Kinase in Aspergillus oryzae [J]. Biotechnology Bulletin, 2023, 39(12): 311-319. |
| [15] | YANG Xu-yan, ZHAO Shuang, MA Tian-yi, BAI Yu, WANG Yu-shu. Cloning of Three Cabbage WRKY Genes and Their Expressions in Response to Abiotic Stress [J]. Biotechnology Bulletin, 2023, 39(11): 261-269. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||