








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (9): 291-300.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0336
Previous Articles Next Articles
ZHANG Man-yu1,2( ), DONG Jia-cheng1,2, GOU Fu-fan1,2, GONG Chao-hui1,2, LIU Qian2, SUN Wen-liang2, KONG zhen3, HAO Jie4, WANG Min1, TIAN Chao-guang2(
), DONG Jia-cheng1,2, GOU Fu-fan1,2, GONG Chao-hui1,2, LIU Qian2, SUN Wen-liang2, KONG zhen3, HAO Jie4, WANG Min1, TIAN Chao-guang2( )
)
Received:2024-04-09
Online:2024-09-26
Published:2024-10-12
Contact:
TIAN Chao-guang
E-mail:zhangmy@tib.cas.cn;tian_cg@tib.cas.cn
ZHANG Man-yu, DONG Jia-cheng, GOU Fu-fan, GONG Chao-hui, LIU Qian, SUN Wen-liang, KONG zhen, HAO Jie, WANG Min, TIAN Chao-guang. Cloning, Expression, Characterization and Application of the Pectin Esterase MtCE12-1 from Myceliophthora thermophila[J]. Biotechnology Bulletin, 2024, 40(9): 291-300.
| 名称 Primer | 序列 DNA sequence(5'-3') | 用途 Application |
|---|---|---|
| Mtce12-1-RT-F | GACGACATTGTAGTGATC | 基因表达分析 |
| Mtce12-1-RT-R | TAGTGGTTGAAGGTGTAG | 基因表达分析 |
| actin-RT-F | AACGCTCCTGCCTTCTAC | 基因表达分析 |
| actin-RT-R | GTAACACCATCACCAGAGTC | 基因表达分析 |
| Mtce12-1-F | GCCAGTTTCGTTCTTCAGAACTAGTATGCGACCGTGGTCGACTCT | 基因克隆 |
| Mtce12-1-R | GTGGTGGTGGTGGTGGTGGATATCGAACACCTGAGGCACCGGGG | 基因克隆 |
| Ptef1-F | CTTCGACCCCTCCTCAAATCTTCTT | 基因鉴定 |
| TtrpC-R | GAGCTATTAAATCACTAGAAGGCAC | 基因鉴定 |
| Mtalp1-ko-F | TTCTGGCCTGCCCTTTTCTTTCAAC | 基因鉴定 |
| Mtalp1-ko-R | GCCCCTTCTTCCGAAAGGGGAGGTA | 基因鉴定 |
Table 1 Primers for amplifying the target sequence
| 名称 Primer | 序列 DNA sequence(5'-3') | 用途 Application |
|---|---|---|
| Mtce12-1-RT-F | GACGACATTGTAGTGATC | 基因表达分析 |
| Mtce12-1-RT-R | TAGTGGTTGAAGGTGTAG | 基因表达分析 |
| actin-RT-F | AACGCTCCTGCCTTCTAC | 基因表达分析 |
| actin-RT-R | GTAACACCATCACCAGAGTC | 基因表达分析 |
| Mtce12-1-F | GCCAGTTTCGTTCTTCAGAACTAGTATGCGACCGTGGTCGACTCT | 基因克隆 |
| Mtce12-1-R | GTGGTGGTGGTGGTGGTGGATATCGAACACCTGAGGCACCGGGG | 基因克隆 |
| Ptef1-F | CTTCGACCCCTCCTCAAATCTTCTT | 基因鉴定 |
| TtrpC-R | GAGCTATTAAATCACTAGAAGGCAC | 基因鉴定 |
| Mtalp1-ko-F | TTCTGGCCTGCCCTTTTCTTTCAAC | 基因鉴定 |
| Mtalp1-ko-R | GCCCCTTCTTCCGAAAGGGGAGGTA | 基因鉴定 |
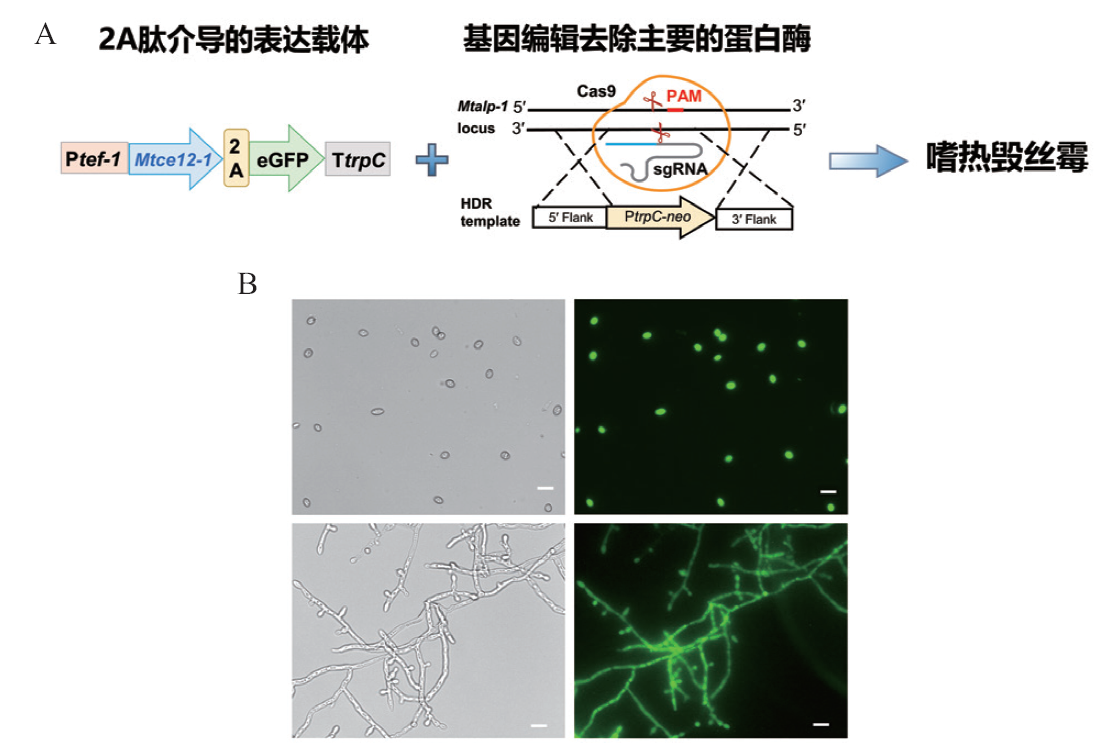
Fig. 2 Recombinant expressed pectin esterase MtCE12-1 in M. thermophile A: Schematic illustration of the expressed vector and recombinant expression of MtCE12-1. B: Microscopic fluorescence imaging of conidia and mycelia of the overexpressing strain OE-MtCE12-1-GFP. Scale bar 10 μm
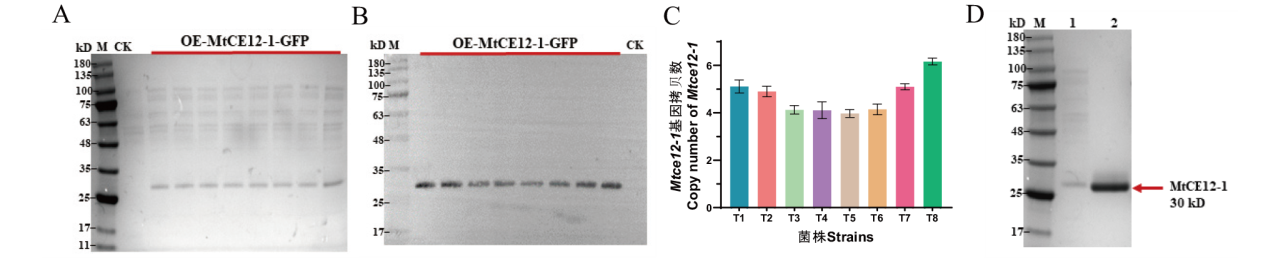
Fig. 3 Detecting the expression of recombinant protein MtCE12-1 by using the SDS-PAGE electrophoresis, Western blotting, and copy number A: SDS-PAGE analysis of secreted proteins from the CK and OE-MtCE12-1-GFP strains after 4-day of growth. M: the protein molecular weight markers. CK is used as the negative control for the transformation of the empty vector pAN52-1N into the host wild-type strain. B: Western blotting of culture supernatants from the strains and probed with anti-His antibody. C: Assay of Mtce12-1 copy number in overexpressing strains by RT-qPCR. D: SDS-PAGE analysis of the purified MtCE12-1. M: the protein molecular weight markers; Lane 1: the culture supernatants of the OE-MtCE12-1-GFP strain T8; Lane 2: the purified enzyme of MtCE12-1
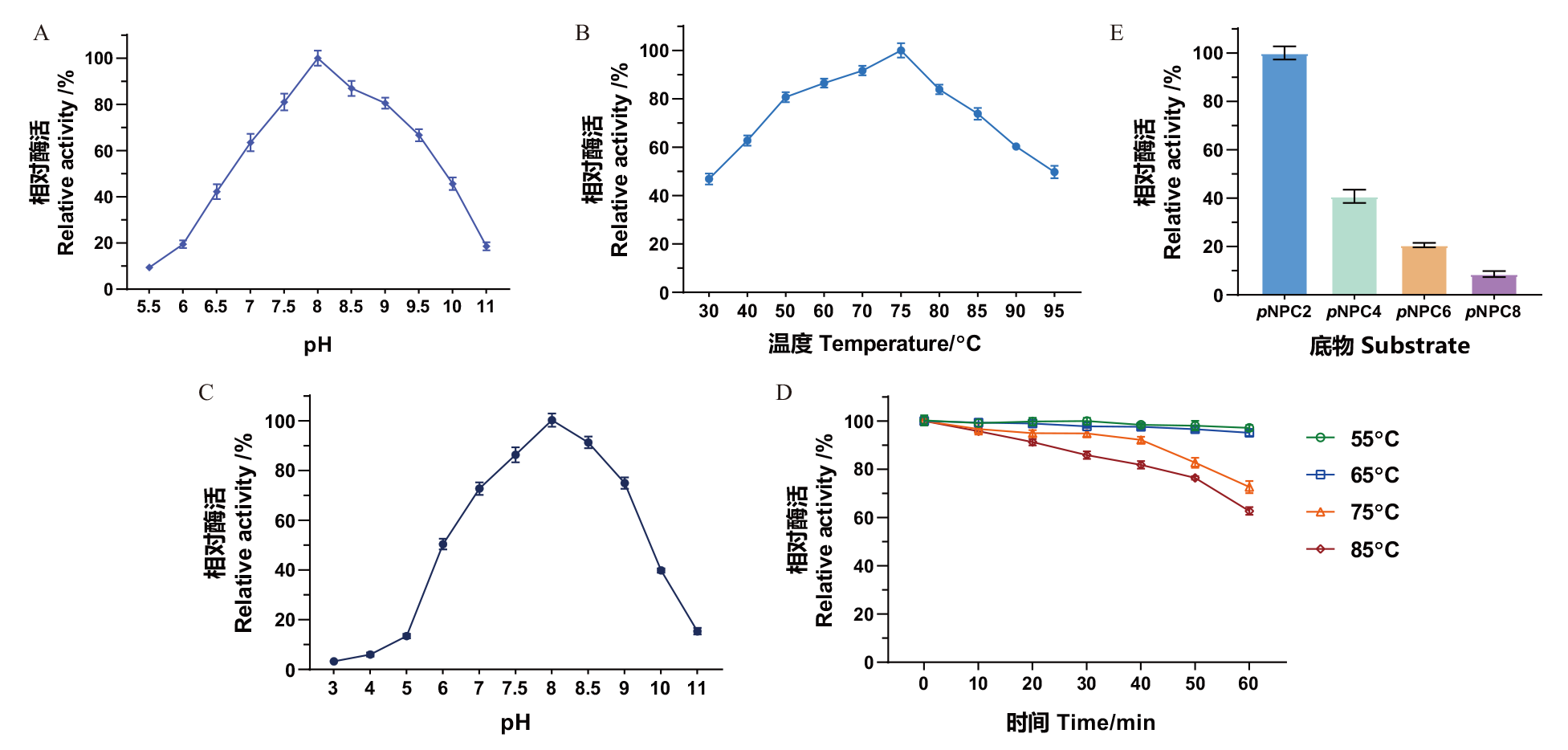
Fig. 4 Enzymatic properties of pectin esterase MtCE12-1 A: Effect of pH on the enzyme activity of MtCE12-1. B: Effect of temperature on the enzyme activity of MtCE12-1. C: Effect of pH on the enzyme activity of MtCE12-1. D: Effect of temperature on the stability of MtCE12-1. E: Substrate specificity of MtCE12-1
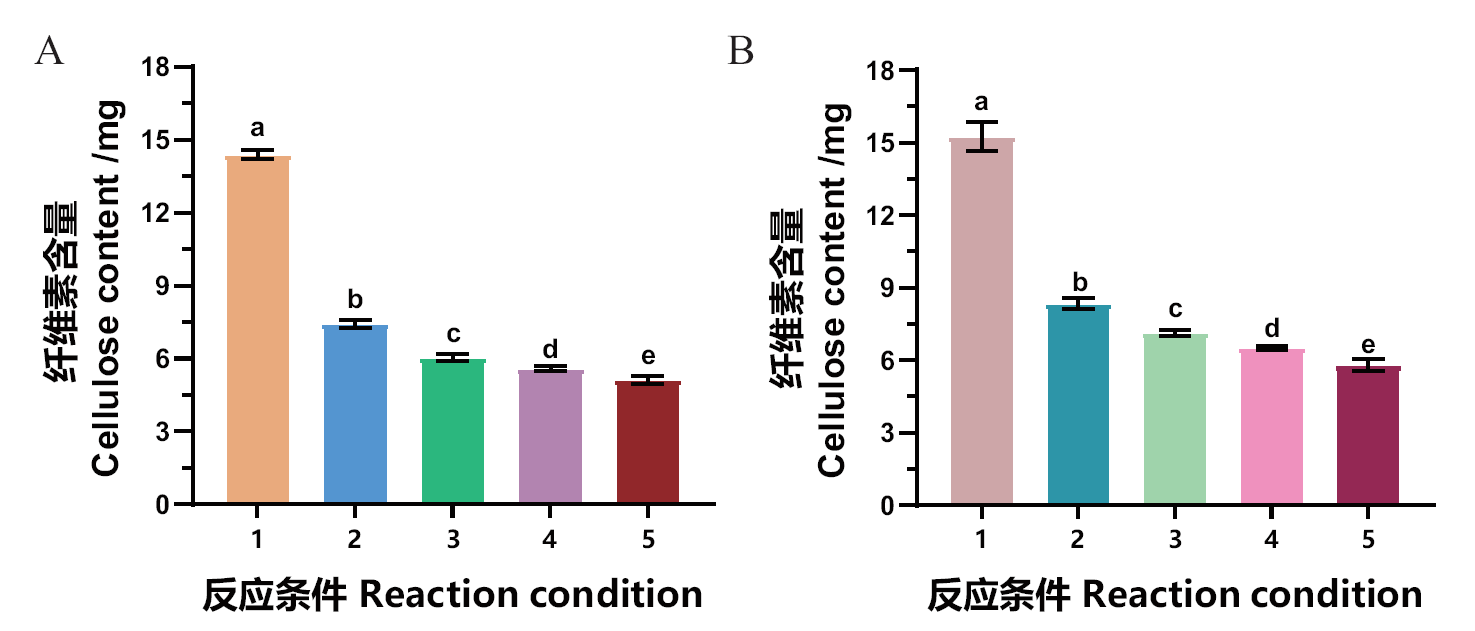
Fig. 5 Synergistic degradation of tobacco waste biomass by pectin esterase MtCE12-1 and cellulase A: Synergistic degradation of tobacco bar by MtCE12-1 and cellulase. B: Synergistic degradation of tobacco stem by MtCE12-1 and cellulase. 1: Control(no enzyme); 2: only cellulase; 3: 100 μg MtCE12-1 and cellulase; 4: 200 μg MtCE12-1 and cellulase; 5: 300 μg MtCE12-1 and cellulase. Different letters are statistically significant(Tukey’s HSD, P < 0.05)
| [1] | Zhang GT, Zhan JJ, Fu HQ. Trends in smoking prevalence and intensity between 2010 and 2018: implications for tobacco control in China[J]. Int J Environ Res Public Health, 2022, 19(2): 670. |
| [2] | Zhang Y, Li RD, Shang GL, et al. Effects of multiscale-mechanical fragmentation on techno-functional properties of industrial tobacco waste[J]. Powder Technol, 2022, 402: 117327. |
| [3] | Tao JM, Chen QS, Chen SY, et al. Metagenomic insight into the microbial degradation of organic compounds in fermented plant leaves[J]. Environ Res, 2022, 214(Pt 1): 113902. |
| [4] | Lin YN, Wang C, Yu GF, et al. Transformation of tobacco biomass into value-added carbohydrate, aromatics, and biochar[J]. Biomass Convers Biorefin, 2024, 14(10): 11697-11705. |
| [5] | 黄申, 芦尧, 刘强, 等. 生物酶在烟草工业中的应用研究进展[J]. 轻工学报, 2023, 38(5): 112-118. |
| Huang S, Lu Y, Liu Q, et al. Review on application of biological enzymes in tobacco industry[J]. J Light Ind, 2023, 38(5): 112-118. | |
| [6] |
郝捷, 李选文, 张宝, 等. 纤维素酶在烟草中的应用进展[J]. 生物技术进展, 2023, 13(2): 166-173.
doi: 10.19586/j.2095-2341.2022.0127 |
|
Hao J, Li XW, Zhang B, et al. Application progress of cellulase in tobacco[J]. Curr Biotechnol, 2023, 13(2): 166-173.
doi: 10.19586/j.2095-2341.2022.0127 |
|
| [7] | Zou G, Shi SH, Jiang YP, et al. Construction of a cellulase hyper-expression system in Trichoderma reesei by promoter and enzyme engineering[J]. Microb Cell Fact, 2012, 11: 21. |
| [8] | Daly P, Cai F, Kubicek CP, et al. From lignocellulose to plastics: knowledge transfer on the degradation approaches by fungi[J]. Biotechnol Adv, 2021, 50: 107770. |
| [9] |
Meng JL, Mäkelä MR, de Vries RP. Molecular engineering to improve lignocellulosic biomass based applications using filamentous fungi[J]. Adv Appl Microbiol, 2021, 114: 73-109.
doi: 10.1016/bs.aambs.2020.09.001 pmid: 33934853 |
| [10] | Kun RS, Gomes ACS, Hildén KS, et al. Developments and opportunities in fungal strain engineering for the production of novel enzymes and enzyme cocktails for plant biomass degradation[J]. Biotechnol Adv, 2019, 37(6): 107361. |
| [11] | Berka RM, Grigoriev IV, Otillar R, et al. Comparative genomic analysis of the thermophilic biomass-degrading fungi Myceliophthora thermophila and Thielavia terrestris[J]. Nat Biotechnol, 2011, 29: 922-927. |
| [12] | Kolbusz MA, Di Falco M, Ishmael N, et al. Transcriptome and exoproteome analysis of utilization of plant-derived biomass by Myceliophthora thermophila[J]. Fungal Genet Biol, 2014, 72: 10-20. |
| [13] |
Liu Q, Li JG, Gao RR, et al. CLR-4, a novel conserved transcription factor for cellulase gene expression in ascomycete fungi[J]. Mol Microbiol, 2019, 111(2): 373-394.
doi: 10.1111/mmi.14160 pmid: 30474279 |
| [14] | Li N, Liu Y, Liu DF, et al. MtTRC-1, a novel transcription factor, regulates cellulase production via directly modulating the genes expression of the Mthac-1 and Mtcbh-1 in Myceliophthora thermophila[J]. Appl Environ Microbiol, 2022, 88(19): e0126322. |
| [15] | Zhu ZJ, Zhang MY, Liu DD, et al. Development of the thermophilic fungus Myceliophthora thermophila into glucoamylase hyperproduction system via the metabolic engineering using improved AsCas12a variants[J]. Microb Cell Fact, 2023, 22(1): 150. |
| [16] | Li JG, Lin LC, Sun T, et al. Direct production of commodity chemicals from lignocellulose using Myceliophthora thermophila[J]. Metab Eng, 2020, 61: 416-426. |
| [17] | Liu J, Chen MX, Gu SY, et al. Independent metabolism of oligosaccharides is the keystone of synchronous utilization of cellulose and hemicellulose in Myceliophthora[J]. PNAS Nexus, 2024, 3(2): pgae053. |
| [18] | Gu SY, Wu TJ, Zhao JQ, et al. Rewiring metabolic flux to simultaneously improve malate production and eliminate by-product succinate accumulation by Myceliophthora thermophila[J]. Microb Biotechnol, 2024, 17(2): e14410. |
| [19] | Kashyap DR, Vohra PK, Chopra S, et al. Applications of pectinases in the commercial sector: a review[J]. Bioresour Technol, 2001, 77(3): 215-227. |
| [20] | Hoondal G, Tiwari R, Tewari R, et al. Microbial alkaline pectinases and their industrial applications: a review[J]. Appl Microbiol Biotechnol, 2002, 59(4): 409-418. |
| [21] |
Lara-Márquez A, Zavala-Páramo MG, López-Romero E, et al. Biotechnological potential of pectinolytic complexes of fungi[J]. Biotechnol Lett, 2011, 33(5): 859-868.
doi: 10.1007/s10529-011-0520-0 pmid: 21246254 |
| [22] | Sista Kameshwar AK, Qin WS. Structural and functional properties of pectin and lignin-carbohydrate complexes de-esterases: a review[J]. Bioresour Bioprocess, 2018, 5(1): 43. |
| [23] |
Schmitz K, Protzko R, Zhang LS, et al. Spotlight on fungal pectin utilization-from phytopathogenicity to molecular recognition and industrial applications[J]. Appl Microbiol Biotechnol, 2019, 103(6): 2507-2524.
doi: 10.1007/s00253-019-09622-4 pmid: 30694345 |
| [24] | Benz JP, Chau BH, Zheng DA, et al. A comparative systems analysis of polysaccharide-elicited responses in Neurospora crassa reveals carbon source-specific cellular adaptations[J]. Mol Microbiol, 2014, 91(2): 275-299. |
| [25] | Liu Q, Zhang YL, Li FY, et al. Upgrading of efficient and scalable CRISPR-Cas-mediated technology for genetic engineering in thermophilic fungus Myceliophthora thermophila[J]. Biotechnol Biofuels, 2019, 12: 293. |
| [26] | Yang YJ, Liu Y, Liu DD, et al. Development of a flow cytometry-based plating-free system for strain engineering in industrial fungi[J]. Appl Microbiol Biotechnol, 2022, 106(2): 713-727. |
| [27] | Liu Q, Gao RR, Li JG, et al. Development of a genome-editing CRISPR/Cas9 system in thermophilic fungal Myceliophthora species and its application to hyper-cellulase production strain engineering[J]. Biotechnol Biofuels, 2017, 10: 1. |
| [28] | Gao LW, Liu GD, Zhao QQ, et al. Customized optimization of lignocellulolytic enzyme cocktails for efficient conversion of pectin-rich biomass residues[J]. Carbohydr Polym, 2022, 297: 120025. |
| [29] |
Sakai T, Sakamoto T, Hallaert J, et al. Pectin, pectinase and protopectinase: production, properties, and applications[J]. Adv Appl Microbiol, 1993, 39: 213-294.
pmid: 8213306 |
| [30] | 华婷, 李雅楠, 王凯凯, 等. 蓝状菌(Talaromyces leycettanus JCM12802)高温果胶甲酯酶PmeT在毕赤酵母中的高效表达及酶学性质[J]. 微生物学报, 2018, 58(1): 122-130. |
| Hua T, Li YN, Wang KK, et al. High-level expression and characterization of pectin methylesterase Pme T from Talaromyces leycettanus JCM12802 in Pichia pastoris[J]. Acta Microbiol Sin, 2018, 58(1): 122-130. | |
| [31] | Kumar R, Meghwanshi GK, Marcianò D, et al. Sequence, structure and functionality of pectin methylesterases and their use in sustainable carbohydrate bioproducts: a review[J]. Int J Biol Macromol, 2023, 244: 125385. |
| [1] | WANG Qian, ZHOU Jia-yan, WANG Qian, DENG Yu-ping, ZHANG Min-hui, CHEN Jing, YANG Jun, ZOU Jian. Identification and Expression Analysis of the YABBY Gene Family in Sunflower [J]. Biotechnology Bulletin, 2024, 40(8): 199-211. |
| [2] | YANG Wei, ZHAO Li-fen, TANG Bing, ZHOU Lin-bi, YANG Juan, MO Chuan-yuan, ZHANG Bao-hui, LI Fei, RUAN Song-lin, DENG Ying. Genome-wide Identification and Expression Analysis of the SRO Gene Family in Brassica juncea L. [J]. Biotechnology Bulletin, 2024, 40(8): 129-141. |
| [3] | HU Ya-dan, WU Guo-qiang, LIU Chen, WEI Ming. Roles of MYB Transcription Factor in Regulating the Responses of Plants to Stress [J]. Biotechnology Bulletin, 2024, 40(6): 5-22. |
| [4] | QIN Jian, LI Zhen-yue, HE Lang, LI Jun-ling, ZHANG Hao, DU Rong. Change of Single-cell Transcription Profile and Analysis of Intercellular Communication in Myogenic Cell Differentiation [J]. Biotechnology Bulletin, 2024, 40(6): 330-342. |
| [5] | CHEN Chun-lin, LI Bai-xue, LI Jin-ling, DU Qing-jie, LI Meng, XIAO Huai-juan. Identification and Expression Analysis of Epidermal Patterning Factor (EPF) Genes in Cucumis melo [J]. Biotechnology Bulletin, 2024, 40(4): 130-138. |
| [6] | CHEN Qiang, HUANG Xin-hui, ZHANG Zheng, ZHANG Chong, LIU Ye-fei. Effects of Melatonin on the Fruit Softening and Ethylene Synthesis of Post-harvest Oriental Melon [J]. Biotechnology Bulletin, 2024, 40(4): 139-147. |
| [7] | ZHANG Yu, SHI Lei, GONG Lei, NIE Feng-jie, YANG Jiang-wei, LIU Xuan, YANG Wen-jing, ZHANG Guo-hui, XIE Rui-xia, ZHANG Li. Genome-wide Identification of Potato WOX Gene Family and Its Expression Analysis in in vitro Regeneration and Abiotic Stress [J]. Biotechnology Bulletin, 2024, 40(3): 170-180. |
| [8] | WU Xing-xing, HONG Hai-bo, GAN Zhi-cheng, LI Rui-ning, HUANG Xian-zhong. Cloning and Preliminary Functional Analysis of CaPI Gene in Capsicum annuum L. [J]. Biotechnology Bulletin, 2024, 40(3): 193-201. |
| [9] | JIANG Lin-qi, ZHAO Jia-ying, ZHENG Fei-xiong, YAO Xin-yi, LI Xiao-xian, YU Zhen-ming. Identification and Expression Analysis of 14-3-3 Gene Family in Dendrobium officinale [J]. Biotechnology Bulletin, 2024, 40(3): 229-241. |
| [10] | ZHENG Fei, YANG Jun-zhao, NIU Yu-feng, LI Rui-lin, ZHAO Guo-zhu. Characterization and Functional Analysis of Lytic Polysaccharide Monooxygenase TtLPMO9I from Thermothelomyces thermophilus [J]. Biotechnology Bulletin, 2024, 40(2): 289-299. |
| [11] | WANG Feng-ting, ZHAO Fu-shun, QIAO Kai-bin, XU Xun, LIU Jin-liang. Progress on the Molecular Mechanism of Scion-rootstock Interactions in Vegetable Grafting [J]. Biotechnology Bulletin, 2024, 40(10): 149-159. |
| [12] | WU Zhen, ZHANG Ming-Ying, YAN Feng, LI Yi-min, GAO Jing, YAN Yong-Gang, ZHANG Gang. Identification and Analysis of WRKY Gene Family in Rheum palmatum L. [J]. Biotechnology Bulletin, 2024, 40(1): 250-261. |
| [13] | YANG Zhi-xiao, HOU Qian, LIU Guo-quan, LU Zhi-gang, CAO Yi, GOU Jian-yu, WANG Yi, LIN Ying-chao. Responses of Rubisco and Rubisco Activase in Different Resistant Tobacco Strains to Brown Spot Stress [J]. Biotechnology Bulletin, 2023, 39(9): 202-212. |
| [14] | CHEN Zhong-yuan, WANG Yu-hong, DAI Wei-jun, ZHANG Yan-min, YE Qian, LIU Xu-ping, TAN Wen-Song, ZHAO Liang. Mechanism Investigation of Ferric Ammonium Citrate on Transfection for Suspended HEK293 Cells [J]. Biotechnology Bulletin, 2023, 39(9): 311-318. |
| [15] | LI Zhi-qi, YUAN Yue, MIAO Rong-qing, PANG Qiu-ying, ZHANG Ai-qin. Melatonin Contents in Eutrema salsugineum and Arabidopsis thaliana Under Salt Stress, and Expression Pattern Analysis of Synthesis Related Genes [J]. Biotechnology Bulletin, 2023, 39(5): 142-151. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||