








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (12): 156-167.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0307
Previous Articles Next Articles
WANG Ya-ping1( ), JIN Lan1, HAO Jin-feng2, CHANG Ming1, WANG Yan-dan1, GAO Feng1(
), JIN Lan1, HAO Jin-feng2, CHANG Ming1, WANG Yan-dan1, GAO Feng1( )
)
Received:2025-03-21
Online:2025-12-26
Published:2026-01-06
Contact:
GAO Feng
E-mail:imypwang@163.com;imgaofeng@163.com
WANG Ya-ping, JIN Lan, HAO Jin-feng, CHANG Ming, WANG Yan-dan, GAO Feng. Identification and Expression Characteristics Analysis of CmRGLG Gene Family in Melon[J]. Biotechnology Bulletin, 2025, 41(12): 156-167.
引物名称 Primer name | 正向引物序列 Forward primer sequence (5′‒3′) | 反向引物序列 Reverse primer sequence (5′‒3′) | 产物大小 Product size (bp) |
|---|---|---|---|
| CmGAPDH | ATCATTCCTAGCAGCACTGG | TTGGCATCAAATATGCTTGACCTG | 278 |
| CmRGLG1 | AGTGGGGTCATTATGGTTATCCG | CCTCTTTCTTGTCTCAGGGGCT | 116 |
| CmRGLG2 | GGTCAATCCCCCCCTTAT | GCTCCTGTTACCTCCTCCAA | 221 |
| CmRGLG3 | CTTGTTTCGGTTTTGGTGAT | CCAGGTGGTGTTTTAGGATTTC | 262 |
| CmRGLG4 | ATATCTCCCCAGAGAGTTGCA | AAGACCAGGTAGGCTTTGAAT | 97 |
| CmRGLG5 | CAAACTCCACAACAACCACGG | GAGATTAGATGACTCAAGTCCAGCG | 123 |
| CmRGLG6 | CGATTCTCCTAATCCCTACCA | TTGCCCACCACTCTTCTCTA | 280 |
Table 1 RT-qPCR primer sequences
引物名称 Primer name | 正向引物序列 Forward primer sequence (5′‒3′) | 反向引物序列 Reverse primer sequence (5′‒3′) | 产物大小 Product size (bp) |
|---|---|---|---|
| CmGAPDH | ATCATTCCTAGCAGCACTGG | TTGGCATCAAATATGCTTGACCTG | 278 |
| CmRGLG1 | AGTGGGGTCATTATGGTTATCCG | CCTCTTTCTTGTCTCAGGGGCT | 116 |
| CmRGLG2 | GGTCAATCCCCCCCTTAT | GCTCCTGTTACCTCCTCCAA | 221 |
| CmRGLG3 | CTTGTTTCGGTTTTGGTGAT | CCAGGTGGTGTTTTAGGATTTC | 262 |
| CmRGLG4 | ATATCTCCCCAGAGAGTTGCA | AAGACCAGGTAGGCTTTGAAT | 97 |
| CmRGLG5 | CAAACTCCACAACAACCACGG | GAGATTAGATGACTCAAGTCCAGCG | 123 |
| CmRGLG6 | CGATTCTCCTAATCCCTACCA | TTGCCCACCACTCTTCTCTA | 280 |

Fig. 1 Gene structure and conservative motif analysis of CmRGLGs in melonA: Gene structure. B: Conserved motifs. C: Motif sequence logo (The height of each letter indicates the frequency of amino acid occurrence at that position)
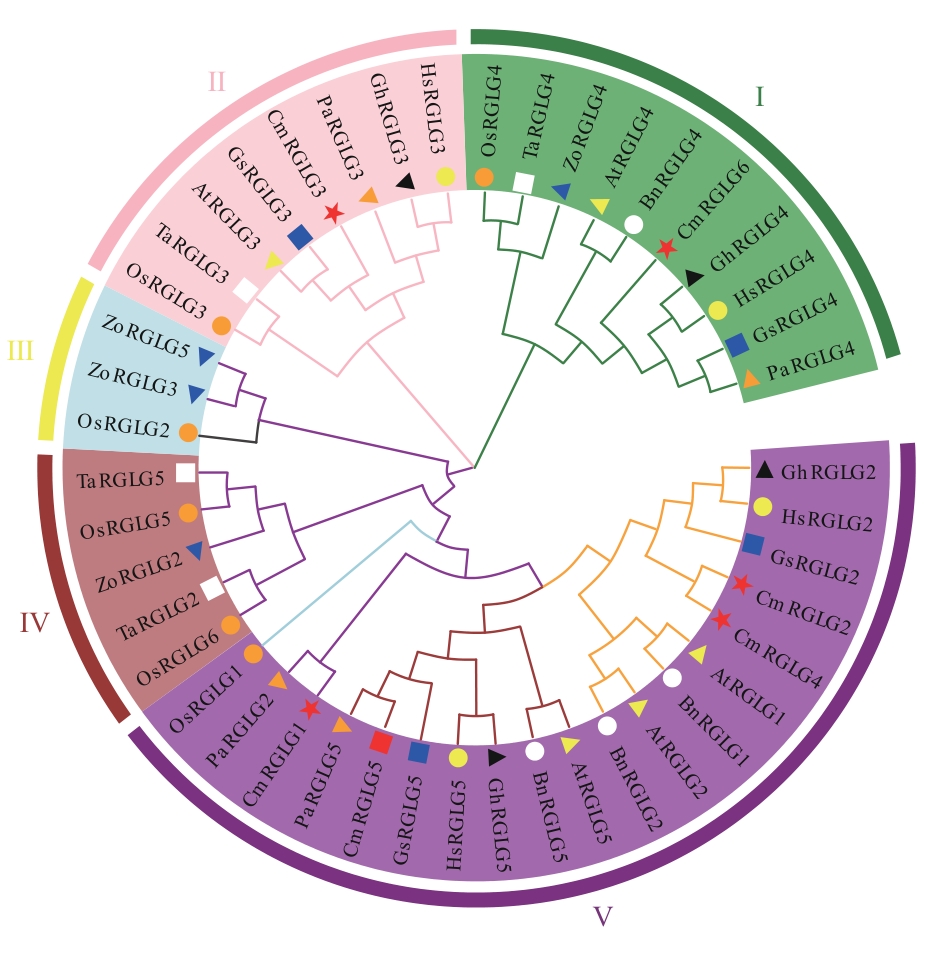
Fig. 2 Phylogenetic tree of RGLG protein family in melon and other speciesCm: Cucumis melo; At: Arabidopsis thaliana; Os: Oryza sativa; Pa: Populus alba; Ta: Triticum aestivum; Gs: Glycine soja; Gh: Gossypium hirsutum; Bn: Brassica napus; Hs: Hibiscus syriacus; Zo: Zingiber officinale
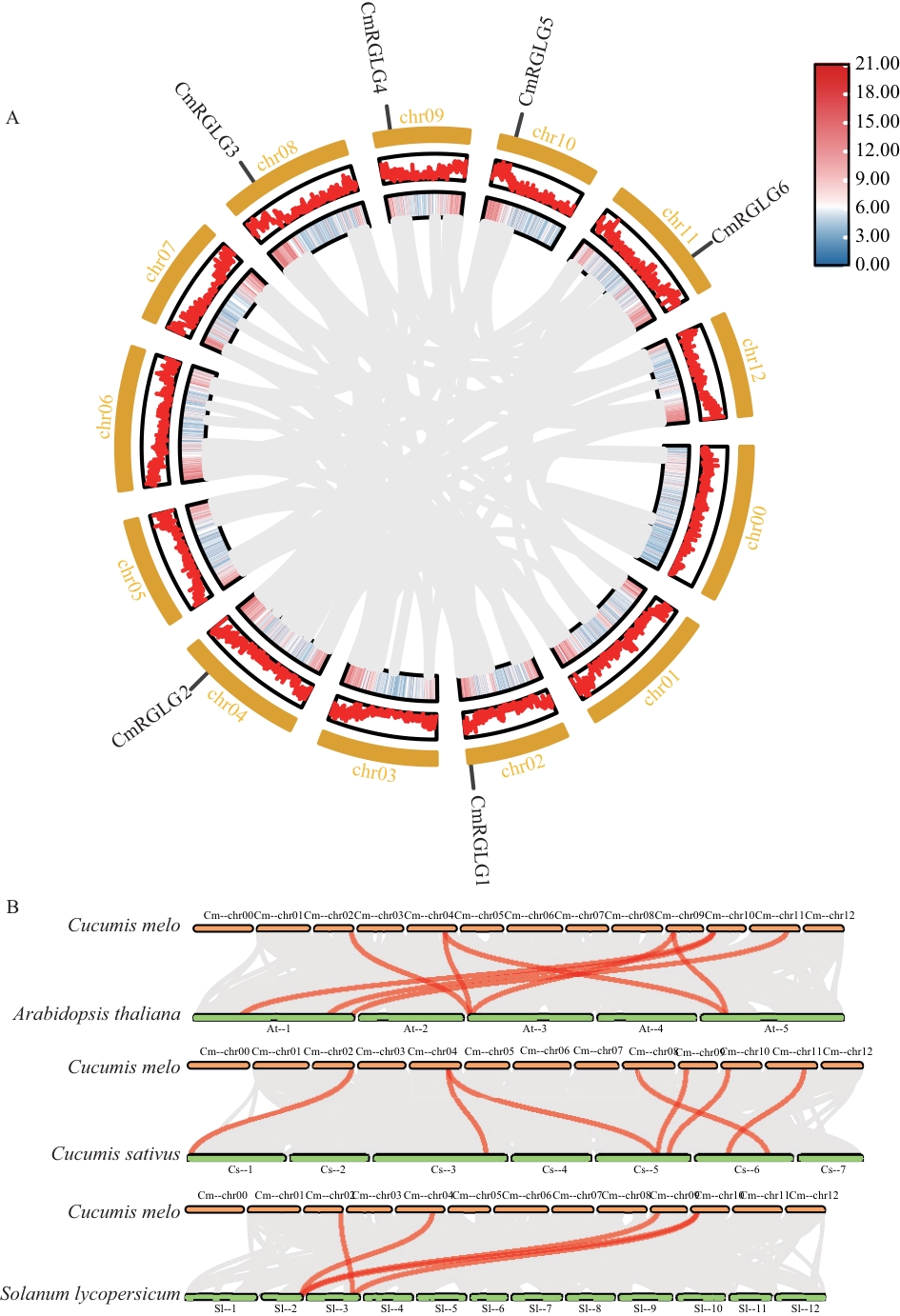
Fig. 4 Intraspecific (A) and interspecific synteny (B) analysis of CmRGLGs in melonGray lines indicate collinear blocks between the melon genome and the genomes of the analyzed species; red lines highlight RGLG gene pairs
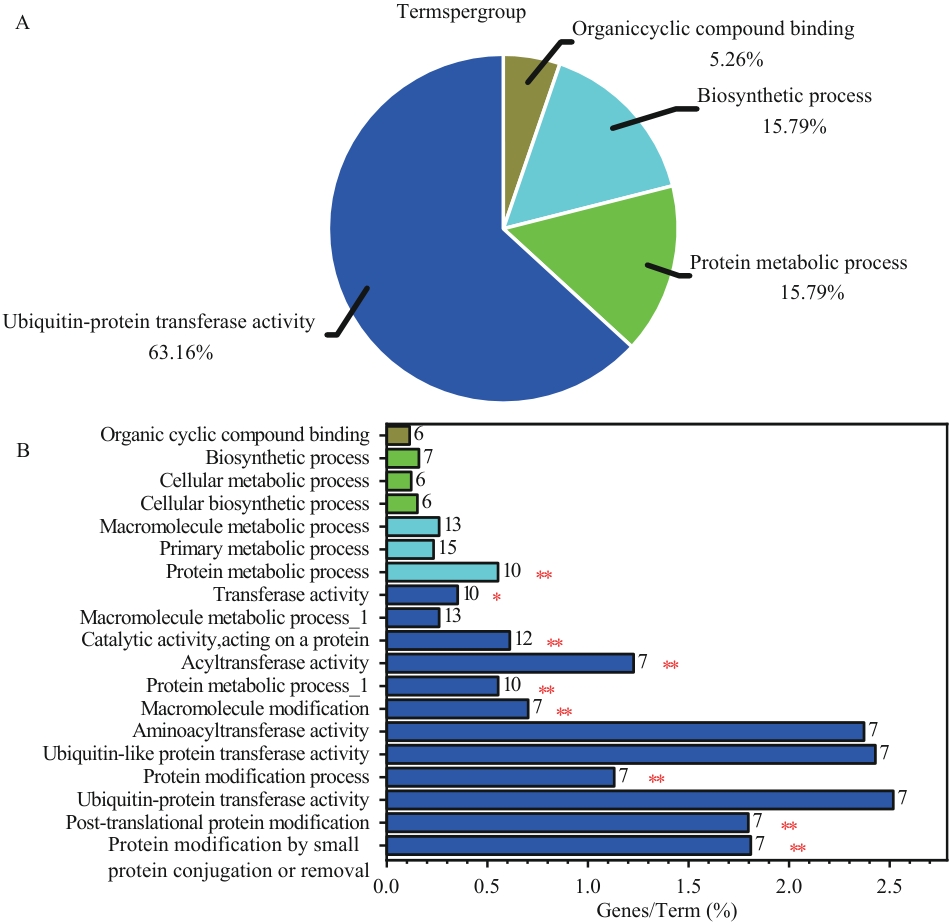
Fig. 6 Statistical results of KEGG and GO annotation in protein-protein interaction analysis by Cytoscape 3.10.2A: KEGG pathway enrichment analysis. B: GO functional enrichment analysis (X-axis: percentage of enriched genes per Term; numbers at the end of bars indicate the specific gene count in each Term; Y-axis: significantly enriched GO Terms). *P<0.05, **P<0.01
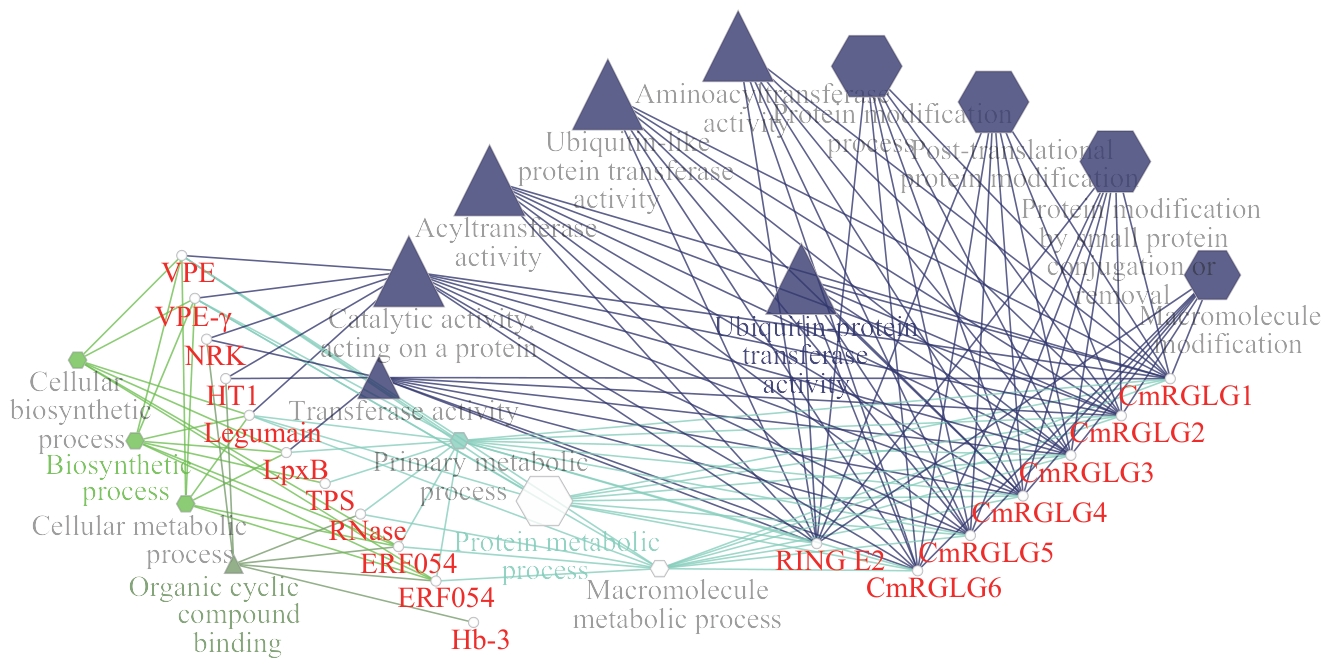
Fig. 7 Protein-protein interaction network of CmRGLGs in melonHexagons indicate biological processes; triangles indicate molecular functions; circles indicate proteins
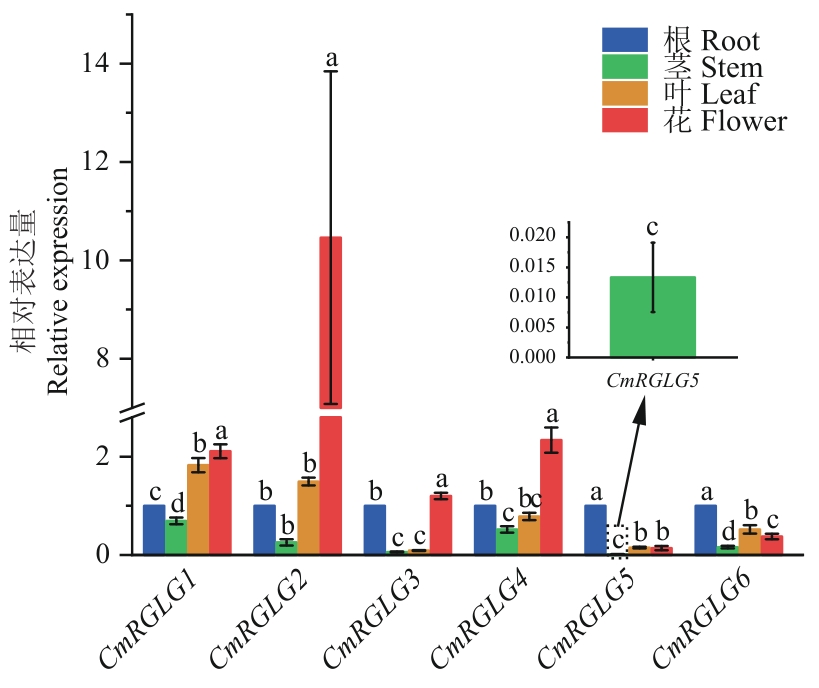
Fig. 8 Analysis of expression characteristics of CmRGLGs in different organs of melonDifferent lowercase letters indicate significant differences (P<0.05). The same below
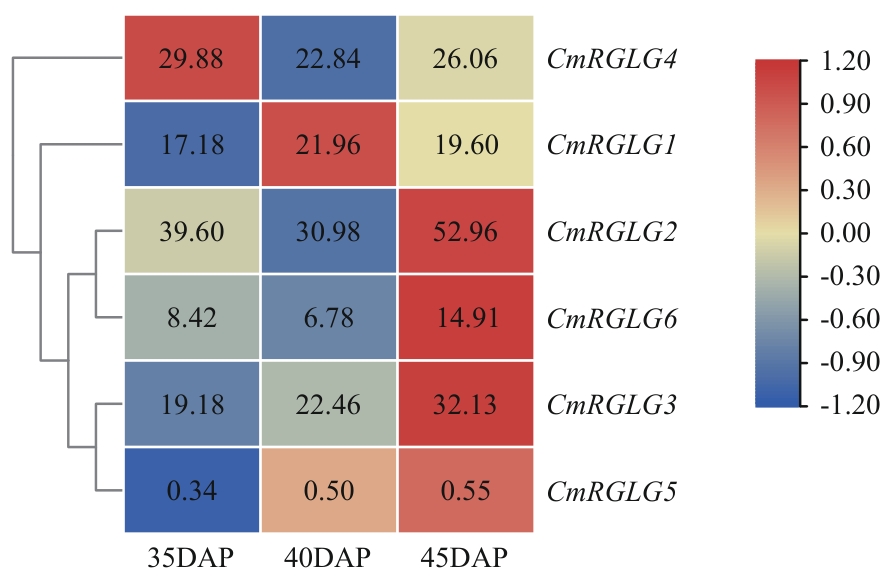
Fig. 10 Analysis of expression characteristics of CmRGLGs in the transcriptome data of melon fruits35, 40, and 45 DAP indicate 35, 40, and 45 d after pollination (DAP), respectively
| [1] | Lobaina DP, Tarazi R, Castorino T, et al. The ubiquitin-proteasome system (UPS) and viral infection in plants [J]. Plants, 2022, 11(19): 2476. |
| [2] | Chen X, Dou QP, Liu JB, et al. Targeting ubiquitin-proteasome system with copper complexes for cancer therapy [J]. Front Mol Biosci, 2021, 8: 649151. |
| [3] | Yang Q, Zhao JY, Chen D, et al. E3 ubiquitin ligases: styles, structures and functions [J]. Mol Biomed, 2021, 2(1): 23. |
| [4] | Lee JH, Kim WT. Regulation of abiotic stress signal transduction by E3 ubiquitin ligases in Arabidopsis [J]. Mol Cells, 2011, 31(3): 201-208. |
| [5] | Du Z, Zhou X, Li L, et al. plantsUPS: a database of plants’ ubiquitin proteasome system [J]. BMC Genomics, 2009, 10: 227. |
| [6] | Deshaies RJ, Joazeiro CAP. RING domain E3 ubiquitin ligases [J]. Annu Rev Biochem, 2009, 78: 399-434. |
| [7] | Saxena H, Negi H, Sharma B. Role of F-box E3-ubiquitin ligases in plant development and stress responses [J]. Plant Cell Rep, 2023, 42(7): 1133-1146. |
| [8] | Sharma B, Saxena H, Negi H. Genome-wide analysis of HECT E3 ubiquitin ligase gene family in Solanum lycopersicum [J]. Sci Rep, 2021, 11(1): 15891. |
| [9] | 苌兴超, 王雪松, 张沿政, 等. 大豆GmPUB32基因的克隆及耐盐功能分析 [J]. 中国油料作物学报, 2021, 43(4): 638-647. |
| Chang XC, Wang XS, Zhang YZ, et al. Cloning and salt resistance analysis of soybean GmPUB32 gene [J]. Chin J Oil Crop Sci, 2021, 43(4): 638-647. | |
| [10] | 徐焕焕, 张芳毓, 刘茜, 等. 番茄SlSL1重组蛋白的表达、纯化及体外泛素化活性检测 [J]. 合肥工业大学学报: 自然科学版, 2022, 45(8): 1130-1134. |
| Xu HH, Zhang FY, Liu Q, et al. Expression, purification and in vitro ubiquitination analysis of tomato SlSL1 recombinant protein [J]. J Hefei Univ Technol Nat Sci, 2022, 45(8): 1130-1134. | |
| [11] | Freemont PS. Ubiquitination: RING for destruction [J]. Curr Biol, 2000, 10(2): R84-R87. |
| [12] | Pan IC, Tsai HH, Cheng YT, et al. Post-transcriptional coordination of the Arabidopsis iron deficiency response is partially dependent on the E3 ligases RING domain ligase1 (RGLG1) and RING domain ligase2 (RGLG2) [J]. Mol Cell Proteom, 2015, 14(10): 2733-2752. |
| [13] | Stone SL, Hauksdóttir H, Troy A, et al. Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis [J]. Plant Physiol, 2005, 137(1): 13-30. |
| [14] | Katoh S, Hong C, Tsunoda Y, et al. High precision NMR structure and function of the RING-H2 finger domain of EL5, a rice protein whose expression is increased upon exposure to pathogen-derived oligosaccharides [J]. J Biol Chem, 2003, 278(17): 15341-15348. |
| [15] | Li YZ, Wu BJ, Yu YL, et al. Genome-wide analysis of the RING finger gene family in apple [J]. Mol Genet Genomics, 2011, 286(1): 81-94. |
| [16] | Qi ZY, Ahammed GJ, Jiang CY, et al. The E3 ubiquitin ligase gene SlRING1 is essential for plant tolerance to cadmium stress in Solanum lycopersicum [J]. J Biotechnol, 2020, 324: 239-247. |
| [17] | Tan B, Lian XD, Cheng J, et al. Genome-wide identification and transcriptome profiling reveal that E3 ubiquitin ligase genes relevant to ethylene, auxin and abscisic acid are differentially expressed in the fruits of melting flesh and stony hard peach varieties [J]. BMC Genomics, 2019, 20(1): 892. |
| [18] | Tang XL, Mei YY, He KX, et al. The RING-type E3 ligase RIE1 sustains leaf longevity by specifically targeting AtACS7 to fine-tune ethylene production in Arabidopsis [J]. Proc Natl Acad Sci USA, 2024, 121(48): e2411271121. |
| [19] | Xin TX, Zhang Z, Li S, et al. Genetic regulation of ethylene dosage for cucumber fruit elongation [J]. Plant Cell, 2019, 31(5): 1063-1076. |
| [20] | Wu T, Wang Y, Jin J, et al. Soybean RING-type E3 ligase GmCHYR16 ubiquitinates the GmERF71 transcription factor for degradation to negatively regulate bicarbonate stress tolerance [J]. New Phytol, 2025, 246(3): 1128-1146. |
| [21] | Shahwar D, Khan Z, Park Y. Molecular marker-assisted mapping, candidate gene identification, and breeding in melon (Cucumis melo L.): a review [J]. Int J Mol Sci, 2023, 24(20): 15490. |
| [22] | Liu YW, Gao YY, Chen MX, et al. GIFTdb: a useful gene database for plant fruit traits improving [J]. Plant J, 2023, 116(4): 1030-1040. |
| [23] | Li ML, Dong XY, Long GZ, et al. Genome-wide analysis of Q-type C2H2 ZFP genes in response to biotic and abiotic stresses in sugar beet [J]. Biology, 2023, 12(10): 1309. |
| [24] | Nystrom SL, McKay DJ. Memes: A motif analysis environment in R using tools from the MEME Suite [J]. PLoS Comput Biol, 2021, 17(9): e1008991. |
| [25] | Zhao DB, Gao FJ, Guan PY, et al. Identification and analysis of differentially expressed trihelix genes in maize (Zea mays) under abiotic stresses [J]. PeerJ, 2023, 11: e15312. |
| [26] | 洪天澍, 海英, 恩和巴雅尔, 等. 甜瓜CmABCG8基因的表达特性分析 [J]. 生物技术通报, 2022, 38(7): 178-185. |
| Hong TS, Hai Y, En H, et al. Analysis of expression characteristics of CmABCG8 gene in Cucumis melo L [J]. Biotechnol Bull, 2022, 38(7): 178-185. | |
| [27] | Chen Q, Liu RJ, Wu YR, et al. ERAD-related E2 and E3 enzymes modulate the drought response by regulating the stability of PIP2 aquaporins [J]. Plant Cell, 2021, 33(8): 2883-2898. |
| [28] | Hao DD, Jin L, Wen X, et al. The RING E3 ligase SDIR1 destabilizes EBF1/EBF2 and modulates the ethylene response to ambient temperature fluctuations in Arabidopsis [J]. Proc Natl Acad Sci USA, 2021, 118(6): e2024592118. |
| [29] | Wang JY, Wang RY, Fang H, et al. Two VOZ transcription factors link an E3 ligase and an NLR immune receptor to modulate immunity in rice [J]. Mol Plant, 2021, 14(2): 253-266. |
| [30] | Wu Q, Zhang X, Peirats-Llobet M, et al. Ubiquitin ligases RGLG1 and RGLG5 regulate abscisic acid signaling by controlling the turnover of phosphatase PP2CA [J]. Plant Cell, 2016, 28(9): 2178-2196. |
| [31] | 舒合凤, 胡荣美, 朱灵, 等. 水稻泛素连接酶基因RGLG31的同源克隆及表达分析[J/OL]. 分子植物育种, 2025. . |
| Shu HF, Hu RM, Zhu L, et al. Homologous cloning and expression analysis of rice ubiquitin ligase gene RGLG31[J/OL]. Mol Plant Breed, 2025. . | |
| [32] | Ma JF, Wang RB, Zhao HY, et al. Genome-wide characterization of the VQ genes in Triticeae and their functionalization driven by polyploidization and gene duplication events in wheat [J]. Int J Biol Macromol, 2023, 243: 125264. |
| [33] | Xu GX, Guo CC, Shan HY, et al. Divergence of duplicate genes in exon-intron structure [J]. Proc Natl Acad Sci USA, 2012, 109(4): 1187-1192. |
| [34] | Sun WQ, Li MD, Wang JB. Genome-wide identification and characterization of the RCI2 gene family in allotetraploid Brassica napus compared with its diploid progenitors [J]. Int J Mol Sci, 2022, 23(2): 614. |
| [35] | Nagels Durand A, Iñigo S, Ritter A, et al. The Arabidopsis iron-sulfur protein GRXS17 is a target of the ubiquitin E3 ligases RGLG3 and RGLG4 [J]. Plant Cell Physiol, 2016, 57(9): 1801-1813. |
| [36] | Zhang X, Wu Q, An CC. RGLG3 and RGLG4, novel ubiquitin ligases modulating jasmonate signaling [J]. Plant Signal Behav, 2012, 7(12): 1709-1711. |
| [37] | Zhang X, Wu Q, Ren J, et al. Two novel RING-type ubiquitin ligases, RGLG3 and RGLG4, are essential for jasmonate-mediated responses in Arabidopsis [J]. Plant Physiol, 2012, 160(2): 808-822. |
| [38] | 陈佳琳. E3泛素连接酶CsRGLG4与转录因子CsAP2L调控柑橘果实采后绿霉病抗病性的机制研究 [D]. 重庆: 西南大学, 2024. |
| Chen JL. Mechanism of E3 ubiquitin ligase CsRGLG4 and transcription factor CsAP2L in regulating postharvest green mold resistance in citrus fruits [D]. Chongqing: Southwest University, 2024. | |
| [39] | Cheng MC, Hsieh EJ, Chen JH, et al. Arabidopsis RGLG2, functioning as a RING E3 ligase, interacts with AtERF53 and negatively regulates the plant drought stress response [J]. Plant Physiol, 2012, 158(1): 363-375. |
| [40] | Dong CH, Agarwal M, Zhang Y, et al. The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1 [J]. Proc Natl Acad Sci USA, 2006, 103(21): 8281-8286. |
| [1] | WANG Fang, SHAO Hui-ru, LYU Lin-long, ZHAO Dian, HU Zhen, LYU Jian-zhen, JIANG Liang. Establishment of TurboID Proximity Labeling Technology in Plants and Bacteria [J]. Biotechnology Bulletin, 2025, 41(9): 44-53. |
| [2] | CHENG Ting-ting, LIU Jun, WANG Li-li, LIAN Cong-long, WEI Wen-jun, GUO Hui, WU Yao-lin, YANG Jing-fan, LAN Jin-xu, CHEN Sui-qing. Genome-wide Identification of the Chalcone Isomerase Gene Family in Eucommia ulmoides and Analysis of Their Expression Patterns [J]. Biotechnology Bulletin, 2025, 41(9): 242-255. |
| [3] | XU Xiao-ping, YANG Cheng-long, HE Xing, GUO Wen-jie, WU Jian, FANG Shao-zhong. Cloning of the LoAPS1 and Its Function Analysis during the Process of Dormancy Release in Lilium [J]. Biotechnology Bulletin, 2025, 41(9): 195-206. |
| [4] | DONG Xiang-xiang, MIAO Bai-ling, XU He-juan, CHEN Juan-juan, LI Liang-jie, GONG Shou-fu, ZHU Qing-song. Bioinformatics Analysis and Flowering Regulation Function of FveBBX20 Gene in Woodland Strawberry [J]. Biotechnology Bulletin, 2025, 41(9): 115-123. |
| [5] | LI Shan, MA Deng-hui, MA Hong-yi, YAO Wen-kong, YIN Xiao. Identification and Expression Analysis of SKP1 Gene Family in Grapevine (Vitis vinifera L.) [J]. Biotechnology Bulletin, 2025, 41(9): 147-158. |
| [6] | HUANG Guo-dong, DENG Yu-xing, CHENG Hong-wei, DAN Yan-nan, ZHOU Hui-wen, WU Lan-hua. Genome-wide Identification and Expression Analysis of the ZIP Gene Family in Soybean [J]. Biotechnology Bulletin, 2025, 41(9): 71-81. |
| [7] | GONG Hui-ling, XING Yu-jie, MA Jun-xian, CAI Xia, FENG Zai-ping. Identification of Laccase (LAC) Gene Family in Potato (Solanum tuberosum L.) and Its Expression Analysis under Salt Stresses [J]. Biotechnology Bulletin, 2025, 41(9): 82-93. |
| [8] | LIU Jia-li, SONG Jing-rong, ZHAO Wen-yu, ZHANG Xin-yuan, ZHAO Zi-yang, CAO Yi-bo, ZHANG Ling-yun. Identification of the R2R3-MYB Gene and Expression Analysis of Flavonoid Regulatory Genes in Blueberry [J]. Biotechnology Bulletin, 2025, 41(9): 124-138. |
| [9] | HUA Wen-ping, LIU Fei, HAO Jia-xin, CHEN Chen. Identification and Expression Patterns Analysis of ADH Gene Family in Salvia miltiorrhiza [J]. Biotechnology Bulletin, 2025, 41(8): 211-219. |
| [10] | LA Gui-xiao, ZHAO Yu-long, DAI Dan-dan, YU Yong-liang, GUO Hong-xia, SHI Gui-xia, JIA Hui, YANG Tie-gang. Identification of Plasma Membrane H+-ATPase Gene Family in Safflower and Expression Analysis in Response to Low Nitrogen and Low Phosphorus Stress [J]. Biotechnology Bulletin, 2025, 41(8): 220-233. |
| [11] | CHENG Xue, FU Ying, CHAI Xiao-jiao, WANG Hong-yan, DENG Xin. Identification of LHC Gene Family in Setaria italica and Expression Analysis under Abiotic Stresses [J]. Biotechnology Bulletin, 2025, 41(8): 102-114. |
| [12] | BAI Yu-guo, LI Wan-di, LIANG Jian-ping, SHI Zhi-yong, LU Geng-long, LIU Hong-jun, NIU Jing-ping. Growth-promoting Mechanism of Trichoderma harzianum T9131 on Astragalus membranaceus Seedlings [J]. Biotechnology Bulletin, 2025, 41(8): 175-185. |
| [13] | LAI Shi-yu, LIANG Qiao-lan, WEI Lie-xin, NIU Er-bo, CHEN Ying-e, ZHOU Xin, YANG Si-zheng, WANG Bo. The Role of NbJAZ3 in the Infection of Nicotiana benthamiana by Alfalfa Mosaic Virus [J]. Biotechnology Bulletin, 2025, 41(8): 186-196. |
| [14] | REN Rui-bin, SI Er-jing, WAN Guang-you, WANG Jun-cheng, YAO Li-rong, ZHANG Hong, MA Xiao-le, LI Bao-chun, WANG Hua-jun, MENG Ya-xiong. Identification and Expression Analysis of GH17 Gene Family of Pyrenophora graminea [J]. Biotechnology Bulletin, 2025, 41(8): 146-154. |
| [15] | ZENG Dan, HUANG Yuan, WANG Jian, ZHANG Yan, LIU Qing-xia, GU Rong-hui, SUN Qing-wen, CHEN Hong-yu. Genome-wide Identification and Expression Analysis of bZIP Transcription Factor Family in Dendrobium officinale [J]. Biotechnology Bulletin, 2025, 41(8): 197-210. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||