








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (9): 71-81.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0318
Previous Articles Next Articles
HUANG Guo-dong1( ), DENG Yu-xing1, CHENG Hong-wei2, DAN Yan-nan1, ZHOU Hui-wen1(
), DENG Yu-xing1, CHENG Hong-wei2, DAN Yan-nan1, ZHOU Hui-wen1( ), WU Lan-hua1(
), WU Lan-hua1( )
)
Received:2025-03-26
Online:2025-09-26
Published:2025-09-24
Contact:
ZHOU Hui-wen, WU Lan-hua
E-mail:1444098903@qq.com;zhouhuiwen0320@126.com;wulanhua1990@126.com
HUANG Guo-dong, DENG Yu-xing, CHENG Hong-wei, DAN Yan-nan, ZHOU Hui-wen, WU Lan-hua. Genome-wide Identification and Expression Analysis of the ZIP Gene Family in Soybean[J]. Biotechnology Bulletin, 2025, 41(9): 71-81.
基因 Gene | 上游引物 Forward primer (5'-3') | 下游引物 Reverse primer (5'-3') | 片段大小 Size (bp) |
|---|---|---|---|
| GmZIP6 | GCGTGTTGTGTCCTTCTTTTAC | CTTAACAATGGGAGGCTCACC | 167 |
| GmZIP12 | AGGAGGAATCAGGGGATGTG | GAAGCTCCCATAGAAATTCC | 178 |
| GmZIP20 | TCCTTTTACCAACCATGGCCT | TCAACAATGGGAGGCTCACC | 150 |
| GmZIP23 | TGACCAACCGCGTGATGCTG | GTGATGATGATCAGAGCTAG | 127 |
| GmZIP25 | GAAGTGTAGGACTACTAGTG | GTCTCATGATGAGCATGGTG | 126 |
| Glyma.18G290800(Actin) | GCACCACCGGAGAGAAAATA | GTGCACAATTGATGGACCAG | 123 |
Table 1 Gene primer sequences for real-time PCR
基因 Gene | 上游引物 Forward primer (5'-3') | 下游引物 Reverse primer (5'-3') | 片段大小 Size (bp) |
|---|---|---|---|
| GmZIP6 | GCGTGTTGTGTCCTTCTTTTAC | CTTAACAATGGGAGGCTCACC | 167 |
| GmZIP12 | AGGAGGAATCAGGGGATGTG | GAAGCTCCCATAGAAATTCC | 178 |
| GmZIP20 | TCCTTTTACCAACCATGGCCT | TCAACAATGGGAGGCTCACC | 150 |
| GmZIP23 | TGACCAACCGCGTGATGCTG | GTGATGATGATCAGAGCTAG | 127 |
| GmZIP25 | GAAGTGTAGGACTACTAGTG | GTCTCATGATGAGCATGGTG | 126 |
| Glyma.18G290800(Actin) | GCACCACCGGAGAGAAAATA | GTGCACAATTGATGGACCAG | 123 |
基因 Gene | 基因ID Gene ID | 氨基酸数量 Amino acid | 相对分子质量 Mw(Da) | 等电点 pI | 不稳定系数 Instability index | 脂肪系数 Aliphatic index | 亲水性 Hydropathicity | 亚细胞定位 Subcellular location |
|---|---|---|---|---|---|---|---|---|
| GmZIP1 | Glyma.02G126000 | 360 | 38 468.30 | 7.62 | 26.61 | 104.06 | 0.551 | 细胞膜 |
| GmZIP2 | Glyma.05G137400 | 485 | 51 989.60 | 6.00 | 31.08 | 95.96 | -0.033 | 细胞膜 |
| GmZIP3 | Glyma.06G052000 | 478 | 51 484.51 | 6.57 | 52.00 | 94.14 | 0.340 | 细胞膜 |
| GmZIP4 | Glyma.07G223200 | 356 | 38 455.58 | 8.61 | 33.40 | 108.54 | 0.569 | 细胞膜 |
| GmZIP5 | Glyma.08G092700 | 485 | 52 353.64 | 5.79 | 35.60 | 92.95 | -0.139 | 细胞膜 |
| GmZIP6 | Glyma.08G164400 | 361 | 38 404.85 | 6.38 | 40.99 | 102.16 | 0.446 | 细胞膜 |
| GmZIP7 | Glyma.08G328000 | 349 | 37 559.76 | 6.06 | 32.77 | 107.65 | 0.668 | 细胞膜 |
| GmZIP8 | Glyma.09G271900 | 598 | 62 182.12 | 7.19 | 30.11 | 112.26 | 0.777 | 细胞膜 |
| GmZIP9 | Glyma.11G132500 | 272 | 28 959.34 | 9.10 | 47.79 | 114.38 | 0.693 | 细胞膜 |
| GmZIP10 | Glyma.11G169300 | 326 | 34 776.07 | 5.81 | 29.45 | 113.96 | 0.769 | 液泡膜 |
| GmZIP11 | Glyma.12G056900 | 256 | 27 593.55 | 9.19 | 45.62 | 114.26 | 0.586 | 细胞膜 |
| GmZIP12 | Glyma.13G004400 | 347 | 37 633.21 | 7.72 | 37.42 | 110.55 | 0.455 | 细胞膜 |
| GmZIP13 | Glyma.13G338300 | 350 | 37 989.51 | 8.43 | 30.78 | 105.63 | 0.549 | 细胞膜 |
| GmZIP14 | Glyma.13G340900 | 276 | 29 153.48 | 8.78 | 38.44 | 118.04 | 0.740 | 细胞膜 |
| GmZIP15 | Glyma.14G094902 | 135 | 14 389.27 | 6.50 | 43.26 | 120.00 | 1.076 | 液泡膜 |
| GmZIP16 | Glyma.14G196200 | 324 | 33 863.86 | 6.35 | 34.60 | 114.51 | 0.773 | 细胞膜 |
| GmZIP17 | Glyma.15G033500 | 276 | 29 241.54 | 7.95 | 37.60 | 117.68 | 0.717 | 细胞膜 |
| GmZIP18 | Glyma.15G036200 | 345 | 37 385.49 | 6.82 | 32.57 | 101.22 | 0.462 | 细胞膜 |
| GmZIP19 | Glyma.15G036300 | 342 | 36 681.97 | 8.18 | 31.29 | 106.08 | 0.617 | 细胞膜 |
| GmZIP20 | Glyma.15G262800 | 359 | 38 243.55 | 6.14 | 39.12 | 104.09 | 0.486 | 细胞膜 |
| GmZIP21 | Glyma.17G228600 | 393 | 43 130.94 | 5.73 | 40.52 | 98.52 | 0.475 | 细胞膜 |
| GmZIP22 | Glyma.18G060300 | 328 | 35 036.37 | 5.96 | 29.37 | 112.07 | 0.742 | 细胞膜 |
| GmZIP23 | Glyma.18G078600 | 360 | 38 721.95 | 6.08 | 35.02 | 108.97 | 0.577 | 细胞膜 |
| GmZIP24 | Glyma.18G217100 | 598 | 62 115.03 | 6.93 | 29.44 | 112.59 | 0.781 | 细胞膜 |
| GmZIP25 | Glyma.20G022500 | 358 | 38 705.73 | 6.86 | 34.80 | 105.20 | 0.548 | 细胞膜 |
| GmZIP26 | Glyma.20G063100 | 354 | 38 301.74 | 6.30 | 28.63 | 105.56 | 0.399 | 细胞膜 |
Table 2 Basic information on physicochemical properties of GmZIP gene family members
基因 Gene | 基因ID Gene ID | 氨基酸数量 Amino acid | 相对分子质量 Mw(Da) | 等电点 pI | 不稳定系数 Instability index | 脂肪系数 Aliphatic index | 亲水性 Hydropathicity | 亚细胞定位 Subcellular location |
|---|---|---|---|---|---|---|---|---|
| GmZIP1 | Glyma.02G126000 | 360 | 38 468.30 | 7.62 | 26.61 | 104.06 | 0.551 | 细胞膜 |
| GmZIP2 | Glyma.05G137400 | 485 | 51 989.60 | 6.00 | 31.08 | 95.96 | -0.033 | 细胞膜 |
| GmZIP3 | Glyma.06G052000 | 478 | 51 484.51 | 6.57 | 52.00 | 94.14 | 0.340 | 细胞膜 |
| GmZIP4 | Glyma.07G223200 | 356 | 38 455.58 | 8.61 | 33.40 | 108.54 | 0.569 | 细胞膜 |
| GmZIP5 | Glyma.08G092700 | 485 | 52 353.64 | 5.79 | 35.60 | 92.95 | -0.139 | 细胞膜 |
| GmZIP6 | Glyma.08G164400 | 361 | 38 404.85 | 6.38 | 40.99 | 102.16 | 0.446 | 细胞膜 |
| GmZIP7 | Glyma.08G328000 | 349 | 37 559.76 | 6.06 | 32.77 | 107.65 | 0.668 | 细胞膜 |
| GmZIP8 | Glyma.09G271900 | 598 | 62 182.12 | 7.19 | 30.11 | 112.26 | 0.777 | 细胞膜 |
| GmZIP9 | Glyma.11G132500 | 272 | 28 959.34 | 9.10 | 47.79 | 114.38 | 0.693 | 细胞膜 |
| GmZIP10 | Glyma.11G169300 | 326 | 34 776.07 | 5.81 | 29.45 | 113.96 | 0.769 | 液泡膜 |
| GmZIP11 | Glyma.12G056900 | 256 | 27 593.55 | 9.19 | 45.62 | 114.26 | 0.586 | 细胞膜 |
| GmZIP12 | Glyma.13G004400 | 347 | 37 633.21 | 7.72 | 37.42 | 110.55 | 0.455 | 细胞膜 |
| GmZIP13 | Glyma.13G338300 | 350 | 37 989.51 | 8.43 | 30.78 | 105.63 | 0.549 | 细胞膜 |
| GmZIP14 | Glyma.13G340900 | 276 | 29 153.48 | 8.78 | 38.44 | 118.04 | 0.740 | 细胞膜 |
| GmZIP15 | Glyma.14G094902 | 135 | 14 389.27 | 6.50 | 43.26 | 120.00 | 1.076 | 液泡膜 |
| GmZIP16 | Glyma.14G196200 | 324 | 33 863.86 | 6.35 | 34.60 | 114.51 | 0.773 | 细胞膜 |
| GmZIP17 | Glyma.15G033500 | 276 | 29 241.54 | 7.95 | 37.60 | 117.68 | 0.717 | 细胞膜 |
| GmZIP18 | Glyma.15G036200 | 345 | 37 385.49 | 6.82 | 32.57 | 101.22 | 0.462 | 细胞膜 |
| GmZIP19 | Glyma.15G036300 | 342 | 36 681.97 | 8.18 | 31.29 | 106.08 | 0.617 | 细胞膜 |
| GmZIP20 | Glyma.15G262800 | 359 | 38 243.55 | 6.14 | 39.12 | 104.09 | 0.486 | 细胞膜 |
| GmZIP21 | Glyma.17G228600 | 393 | 43 130.94 | 5.73 | 40.52 | 98.52 | 0.475 | 细胞膜 |
| GmZIP22 | Glyma.18G060300 | 328 | 35 036.37 | 5.96 | 29.37 | 112.07 | 0.742 | 细胞膜 |
| GmZIP23 | Glyma.18G078600 | 360 | 38 721.95 | 6.08 | 35.02 | 108.97 | 0.577 | 细胞膜 |
| GmZIP24 | Glyma.18G217100 | 598 | 62 115.03 | 6.93 | 29.44 | 112.59 | 0.781 | 细胞膜 |
| GmZIP25 | Glyma.20G022500 | 358 | 38 705.73 | 6.86 | 34.80 | 105.20 | 0.548 | 细胞膜 |
| GmZIP26 | Glyma.20G063100 | 354 | 38 301.74 | 6.30 | 28.63 | 105.56 | 0.399 | 细胞膜 |

Fig. 5 Expression patterns of GmZIP in different tissues of soybeanYoung_leaf refers to new leaf, Flower refer to the period of flowering, One cm pod refers to a pod of 1cm in length, and Pod shell refers to a pod shell with 10 d of seed development
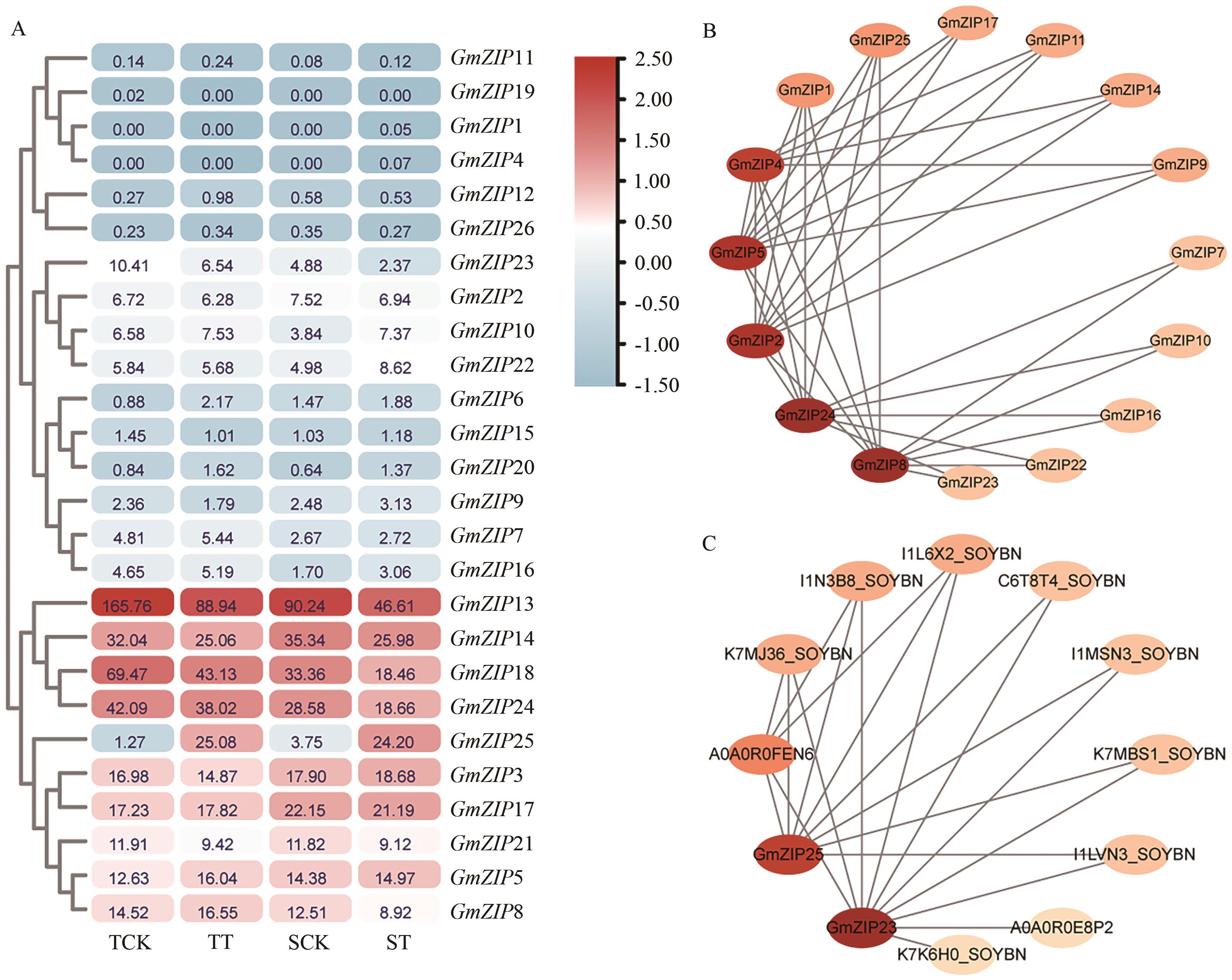
Fig. 6 Expression patterns (A) of GmZIP in response to Al stress and prediction analysis of interacting proteins (B, C) of GmZIPTCK and TT refer to the control group and Al poisoning treated group of soybean Al-tolerant variety Nangong 99-6, while SCK and ST refer to the control group and Al poisoning treated group of soybean Al-sensitive variety Zhongdou 32, respectively
| Gene | TT_T_vs_TCK_T_Log2FC | TT_T_vs_TCK_T_regulated | ST_T_vs_SCK_T_Log2FC | ST_T_vs_SCK_T_regulated |
|---|---|---|---|---|
| GmZIP6 | 1.24 | Up | 0.39 | False |
| GmZIP12 | 1.82 | Up | -0.10 | False |
| GmZIP20 | 0.898 | False | 1.14 | Up |
| GmZIP23 | -0.73 | False | -1.01 | Down |
| GmZIP25 | 4.25 | Up | 2.72 | Up |
Table 3 Differentially expressed GmZIP genes in Al-tolerant and Al-sensitive germplasm
| Gene | TT_T_vs_TCK_T_Log2FC | TT_T_vs_TCK_T_regulated | ST_T_vs_SCK_T_Log2FC | ST_T_vs_SCK_T_regulated |
|---|---|---|---|---|
| GmZIP6 | 1.24 | Up | 0.39 | False |
| GmZIP12 | 1.82 | Up | -0.10 | False |
| GmZIP20 | 0.898 | False | 1.14 | Up |
| GmZIP23 | -0.73 | False | -1.01 | Down |
| GmZIP25 | 4.25 | Up | 2.72 | Up |
| [1] | Ajeesh Krishna TP, Maharajan T, Victor Roch G, et al. Structure, function, regulation and phylogenetic relationship of ZIP family transporters of plants [J]. Front Plant Sci, 2020, 11: 662. |
| [2] | Guerinot ML. The ZIP family of metal transporters [J]. Biochim Biophys Acta BBA Biomembr, 2000, 1465(1-2): 190-198. |
| [3] | Vatansever R, Özyiğit İİ, Filiz E. Comparative and phylogenetic analysis of zinc transporter genes/proteins in plants [J]. Turk J Biol, 2016, 40(3): 600-611. |
| [4] | Grotz N, Fox T, Connolly E, et al. Identification of a family of zinc transporter genes from Arabidopsis that respond to zinc deficiency [J]. Proc Natl Acad Sci USA, 1998, 95(12): 7220-7224. |
| [5] | Zeng X, Yang SX, Li F, et al. Genome-wide identification of OsZIPs in rice and gene expression analysis under manganese and selenium stress [J]. Genes, 2024, 15(6): 696. |
| [6] | Ivanov R, Bauer P. Sequence and coexpression analysis of iron-regulated ZIP transporter genes reveals crossing points between iron acquisition strategies in green algae and land plants [J]. Plant Soil, 2017, 418(1): 61-73. |
| [7] | Mondal TK, Ahmad Ganie S, Rana MK, et al. Erratum to: genome-wide analysis of zinc transporter genes of maize (Zea mays) [J]. Plant Mol Biol Report, 2014, 32(3): 779. |
| [8] | Tang MJ, Zhang XL, Xu L, et al. Genome- and transcriptome-wide characterization of ZIP gene family reveals their potential role in radish (Raphanus sativus) response to heavy metal stresses [J]. Sci Hortic, 2024, 324: 112564. |
| [9] | Nishida S, Tsuzuki C, Kato A, et al. AtIRT1, the primary iron uptake transporter in the root, mediates excess nickel accumulation in Arabidopsis thaliana [J]. Plant Cell Physiol, 2011, 52(8): 1433-1442. |
| [10] | Lin YF, Liang HM, Yang SY, et al. Arabidopsis IRT3 is a zinc-regulated and plasma membrane localized zinc/iron transporter [J]. New Phytol, 2009, 182(2): 392-404. |
| [11] | Lee S, Lee J, Ricachenevsky FK, et al. Redundant roles of four ZIP family members in zinc homeostasis and seed development in Arabidopsis thaliana [J]. Plant J, 2021, 108(4): 1162-1173. |
| [12] | Ochoa Tufiño V, Almira Casellas M, van Duynhoven A, et al. Arabidopsis thaliana Zn transporter genes ZIP3 and ZIP5 provide the main Zn uptake route and act redundantly to face Zn deficiency [J]. Plant J, 2025, 121(3): e17251. |
| [13] | Liu XS, Feng SJ, Zhang BQ, et al. OsZIP1 functions as a metal efflux transporter limiting excess zinc, copper and cadmium accumulation in rice [J]. BMC Plant Biol, 2019, 19(1): 283. |
| [14] | Yang M, Li YT, Liu ZH, et al. A high activity zinc transporter OsZIP9 mediates zinc uptake in rice [J]. Plant J, 2020, 103(5): 1695-1709. |
| [15] | Ishimaru Y, Suzuki M, Kobayashi T, et al. OsZIP4, a novel zinc-regulated zinc transporter in rice [J]. J Exp Bot, 2005, 56(422): 3207-3214. |
| [16] | Ishimaru Y, Masuda H, Suzuki M, et al. Overexpression of the OsZIP4 zinc transporter confers disarrangement of zinc distribution in rice plants [J]. J Exp Bot, 2007, 58(11): 2909-2915. |
| [17] | Kavitha PG, Kuruvilla S, Mathew MK. Functional characterization of a transition metal ion transporter, OsZIP6 from rice (Oryza sativa L.) [J]. Plant Physiol Biochem, 2015, 97: 165-174. |
| [18] | 蔺庆伟, 马剑敏, 彭雪, 等. 环境中铝来源、铝毒机制及影响因子研究进展 [J]. 生态环境学报, 2019, 28(9): 1915-1926. |
| Lin QW, Ma JM, Peng X, et al. Progress on the environmental sources of aluminum, mechanism of aluminum toxicity and its influencing factors [J]. Ecol Environ Sci, 2019, 28(9): 1915-1926. | |
| [19] | Chauhan DK, Yadav V, Vaculík M, et al. Aluminum toxicity and aluminum stress-induced physiological tolerance responses in higher plants [J]. Crit Rev Biotechnol, 2021, 41(5): 715-730. |
| [20] | de Almeida GHG, de Cássia Siqueira-Soares R, Mota TR, et al. Aluminum oxide nanoparticles affect the cell wall structure and lignin composition slightly altering the soybean growth [J]. Plant Physiol Biochem, 2021, 159: 335-346. |
| [21] | 胡湘云, 王奕文, 方幽文, 等. 酸性土壤下缓解大豆铝胁迫的研究进展 [J]. 科学通报, 2023, 68(33): 4517-4531. |
| Hu XY, Wang YW, Fang YW, et al. Research progress on alleviating aluminum stress of soybean in acidic soil [J]. Chin Sci Bull, 2023, 68(33): 4517-4531. | |
| [22] | Li HJ, Wang N, Hu WP, et al. ZmNRAMP4 enhances the tolerance to aluminum stress in Arabidopsis thaliana [J]. Int J Mol Sci, 2022, 23(15): 8162. |
| [23] | Li JY, Liu JP, Dong DK, et al. Natural variation underlies alterations in Nramp aluminum transporter (NRAT1) expression and function that play a key role in rice aluminum tolerance [J]. Proc Natl Acad Sci USA, 2014, 111(17): 6503-6508. |
| [24] | Lu MX, Wang ZG, Fu S, et al. Functional characterization of the SbNrat1 gene in sorghum [J]. Plant Sci, 2017, 262: 18-23. |
| [25] | Rahman R, Upadhyaya H. Aluminium toxicity and its tolerance in plant: a review [J]. J Plant Biol, 2021, 64(2): 101-121. |
| [26] | Zheng X, Chen L, Li XF. Arabidopsis and rice showed a distinct pattern in ZIPs genes expression profile in response to Cd stress [J]. Bot Stud, 2018, 59(1): 22. |
| [27] | Ramesh SA, Shin R, Eide DJ, et al. Differential metal selectivity and gene expression of two zinc transporters from rice [J]. Plant Physiol, 2003, 133(1): 126-134. |
| [28] | Wu X, Zhu ZB, Chen JH, et al. Transcriptome analysis revealed pivotal transporters involved in the reduction of cadmium accumulation in pak choi (Brassica chinensis L.) by exogenous hydrogen-rich water [J]. Chemosphere, 2019, 216: 684-697. |
| [29] | Liu JG, Wang XT, Wang N, et al. Comparative analyses reveal peroxidases play important roles in soybean tolerance to Aluminum toxicity [J]. Agronomy(Basel), 2021, 11(4): 670. |
| [30] | 周会汶, 黄朝平, 汪子怡, 等. 大豆苗期耐铝毒特性综合评价及种质筛选 [J]. 大豆科学, 2022, 41(6): 654-662. |
| Zhou HW, Huang CP, Wang ZY, et al. Comprehensive evaluation of aluminum toxicity tolerance and screening of germplasms during seedling stage in soybean [J]. Soybean Sci, 2022, 41(6): 654-662. | |
| [31] | Chen CJ, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data [J]. Mol Plant, 2020, 13(8): 1194-1202. |
| [32] | Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11 [J]. Mol Biol Evol, 2021, 38(7): 3022-3027. |
| [33] | Ullah A, Shah Z, Munir I, et al. Genome-wide screening and evolutionary analysis of ZIP (ZRT-IRT like proteins) family in cowpea (Vigna unguiculata L.) [J]. Genet Resour Crop Evol, 2024, 71(3): 1145-1157. |
| [34] | Severin AJ, Woody JL, Bolon YT, et al. RNA-seq atlas of Glycine max: a guide to the soybean transcriptome [J]. BMC Plant Biol, 2010, 10: 160. |
| [35] | Zhou HW, Wu LH, Wang RK, et al. Integrated transcriptome and metabolome analysis reveals the response mechanisms of soybean to aluminum toxicity [J]. Plant Soil, 2025. DOI: 10.1007/s11104-024-07151-2 . |
| [36] | 邓兆龙, 孔嘉欣, 李俊营, 等. 普通烟草NtZIP基因家族鉴定及镉胁迫下其表达分析 [J]. 植物生理学报, 2024, 60(7): 1105-1118. |
| Deng ZL, Kong JX, Li JY, et al. Identification of the NtZIP gene family and expression pattern analysis under cadmium stress in tobacco [J]. Plant Physiology Journal, 2024, 60(7): 1105-1118. | |
| [37] | Milner MJ, Seamon J, Craft E, et al. Transport properties of members of the ZIP family in plants and their role in Zn and Mn homeostasis [J]. J Exp Bot, 2013, 64(1): 369-381. |
| [38] | Liu GX, Chen QQ, Li DQ, et al. GmSTOP1-3 increases soybean manganese accumulation under phosphorus deficiency by regulating GmMATE2/13 and GmZIP6/GmIREG3 [J]. Plant Cell Environ, 2025, 48(3): 1812-1828. |
| [39] | Mäser P, Thomine S, Schroeder JI, et al. Phylogenetic relationships within cation transporter families of Arabidopsis [J]. Plant Physiol, 2001, 126(4): 1646-1667. |
| [40] | Moreau S, Thomson RM, Kaiser BN, et al. GmZIP1 encodes a symbiosis-specific zinc transporter in soybean [J]. J Biol Chem, 2002, 277(7): 4738-4746. |
| [1] | CHENG Ting-ting, LIU Jun, WANG Li-li, LIAN Cong-long, WEI Wen-jun, GUO Hui, WU Yao-lin, YANG Jing-fan, LAN Jin-xu, CHEN Sui-qing. Genome-wide Identification of the Chalcone Isomerase Gene Family in Eucommia ulmoides and Analysis of Their Expression Patterns [J]. Biotechnology Bulletin, 2025, 41(9): 242-255. |
| [2] | XU Xiao-ping, YANG Cheng-long, HE Xing, GUO Wen-jie, WU Jian, FANG Shao-zhong. Cloning of the LoAPS1 and Its Function Analysis during the Process of Dormancy Release in Lilium [J]. Biotechnology Bulletin, 2025, 41(9): 195-206. |
| [3] | DONG Xiang-xiang, MIAO Bai-ling, XU He-juan, CHEN Juan-juan, LI Liang-jie, GONG Shou-fu, ZHU Qing-song. Bioinformatics Analysis and Flowering Regulation Function of FveBBX20 Gene in Woodland Strawberry [J]. Biotechnology Bulletin, 2025, 41(9): 115-123. |
| [4] | LI Shan, MA Deng-hui, MA Hong-yi, YAO Wen-kong, YIN Xiao. Identification and Expression Analysis of SKP1 Gene Family in Grapevine (Vitis vinifera L.) [J]. Biotechnology Bulletin, 2025, 41(9): 147-158. |
| [5] | GUAN Zhi-hao, SHAN Zhi-yi, XIONG He, ZHAO Rui-xue. Computational Literature-based Knowledge Discovery for Soybean Coupling Traits [J]. Biotechnology Bulletin, 2025, 41(9): 345-356. |
| [6] | LIU Jia-li, SONG Jing-rong, ZHAO Wen-yu, ZHANG Xin-yuan, ZHAO Zi-yang, CAO Yi-bo, ZHANG Ling-yun. Identification of the R2R3-MYB Gene and Expression Analysis of Flavonoid Regulatory Genes in Blueberry [J]. Biotechnology Bulletin, 2025, 41(9): 124-138. |
| [7] | LI Ya-tao, ZHANG Zhi-peng, ZHAO Meng-yao, LYU Zhen, GAN Tian, WEI Hao, WU Shu-feng, MA Yu-chao. Whole Genome Analysis of Bradyrhizobium sp. Bd1 and the Negative Regulating Function of TetR3 during Cell Growth and Nodulation [J]. Biotechnology Bulletin, 2025, 41(9): 289-301. |
| [8] | LA Gui-xiao, ZHAO Yu-long, DAI Dan-dan, YU Yong-liang, GUO Hong-xia, SHI Gui-xia, JIA Hui, YANG Tie-gang. Identification of Plasma Membrane H+-ATPase Gene Family in Safflower and Expression Analysis in Response to Low Nitrogen and Low Phosphorus Stress [J]. Biotechnology Bulletin, 2025, 41(8): 220-233. |
| [9] | LAI Shi-yu, LIANG Qiao-lan, WEI Lie-xin, NIU Er-bo, CHEN Ying-e, ZHOU Xin, YANG Si-zheng, WANG Bo. The Role of NbJAZ3 in the Infection of Nicotiana benthamiana by Alfalfa Mosaic Virus [J]. Biotechnology Bulletin, 2025, 41(8): 186-196. |
| [10] | ZHU Li-juan, ZHANG Kai, WEN Xiao-lei, CHU Jia-hao, SHI Feng-yu, WANG Yan-li. Mining the Core Genes Being Tolerant to Cadmium in Wild Soybean by WGCNA [J]. Biotechnology Bulletin, 2025, 41(8): 124-136. |
| [11] | ZHAI Ying, JI Jun-jie, CHEN Jiong-xin, YU Hai-wei, LI Shan-shan, ZHAO Yan, MA Tian-yi. Heterologous Overexpression of Soybean GmNF-YB24 Improves the Resistance of Transgenic Tobacco to Drought [J]. Biotechnology Bulletin, 2025, 41(8): 137-145. |
| [12] | NIU Jing-ping, ZHAO Jing, GUO Qian, WANG Shu-hong, ZHAO Jin-zhong, DU Wei-jun, YIN Cong-cong, YUE Ai-qin. Identification and Induced Expression Analysis of Transcription Factors NAC in Soybean Resistance to Soybean Mosaic Virus Based on WGCNA [J]. Biotechnology Bulletin, 2025, 41(7): 95-105. |
| [13] | GONG Yu-han, CHEN Lan, SHANGFANG Hui-zi, HAO Ling-ying, LIU Shuo-qian. Identification and Expression Profile Analysis of the TRB Gene Family in Tea Plant [J]. Biotechnology Bulletin, 2025, 41(7): 214-225. |
| [14] | LI Kai-yue, DENG Xiao-xia, YIN Yuan, DU Ya-tong, XU Yuan-jing, WANG Jing-hong, YU Song, LIN Ji-xiang. Identification of LEA Gene Family and Analysis on Its Response to Aluminum Stress in Ricinus communis L. [J]. Biotechnology Bulletin, 2025, 41(7): 128-138. |
| [15] | ZHAO Qiang, CHEN Si-yu, PENG Fang-li, WANG Can, GAO Jie, ZHOU Ling-bo, ZHANG Guo-bing, JIANG Yu-wen, SHAO Ming-bo. Effects of Intercropping and Nitrogen Application on the Diversity and Functions of Soil Bacteria around Sorghum Rhizosphere [J]. Biotechnology Bulletin, 2025, 41(6): 307-316. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||