








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (8): 267-275.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0388
ZHENG Qian-ming1,2( ), YAN Shuang1,3, XIE Pu1, WANG Hong-lin1,2
), YAN Shuang1,3, XIE Pu1, WANG Hong-lin1,2
Received:2025-04-13
Online:2025-08-26
Published:2025-08-14
Contact:
ZHENG Qian-ming
E-mail:zqm851015@163.com
ZHENG Qian-ming, YAN Shuang, XIE Pu, WANG Hong-lin. Expression and Enzyme Activity Identification of Cell Wall Invertase Gene SmCWIN6 from Red Pitaya[J]. Biotechnology Bulletin, 2025, 41(8): 267-275.
| 名称 Name | 序列 Sequence (5′-3′) |
|---|---|
| CWIN6-qRT-F | GGAATGGGTCTTGATACCTCAG |
| CWIN6-qRT-R | GCAGTGGTTGGATCCCTAAA |
| β-ACT-F | CTTCCATACCAATGAATGAGG |
| β-ACT-R | AACCGCCAAGAGTAGTTCTG |
| CWIN6-GFP-F | ACTAGGGTCTCGCACCATGGGGTTTTATTCTTTGATGATGAT |
| CWIN6-GFP-R | ACTAGGGTCTCTCGCCAGCTATGCTAGCTTTCTTCATGCTCC |
| CWIN6-Y-F | TGGATCCCCCGGGCTGCAGGAATTCATGGGGTTTTATTCTTTGA |
| CWIN6-Y-R | TTGGGTACCGGGCCCCCCCTCGAGCTAAGCTATGCTAGCTTTCT |
Table 1 Primer sequences used in this study
| 名称 Name | 序列 Sequence (5′-3′) |
|---|---|
| CWIN6-qRT-F | GGAATGGGTCTTGATACCTCAG |
| CWIN6-qRT-R | GCAGTGGTTGGATCCCTAAA |
| β-ACT-F | CTTCCATACCAATGAATGAGG |
| β-ACT-R | AACCGCCAAGAGTAGTTCTG |
| CWIN6-GFP-F | ACTAGGGTCTCGCACCATGGGGTTTTATTCTTTGATGATGAT |
| CWIN6-GFP-R | ACTAGGGTCTCTCGCCAGCTATGCTAGCTTTCTTCATGCTCC |
| CWIN6-Y-F | TGGATCCCCCGGGCTGCAGGAATTCATGGGGTTTTATTCTTTGA |
| CWIN6-Y-R | TTGGGTACCGGGCCCCCCCTCGAGCTAAGCTATGCTAGCTTTCT |
名称 Name | 基因ID Gene ID | 定位 Location | 位置 Position (aa) | ||
|---|---|---|---|---|---|
信号肽 Signal peptide | GH32N结构域 GH32N domain | GH32C结构域 GH32C domain | |||
| SmCWIN1 | HU02G02988.1 | 细胞壁Cell wall | 1-19 | 48-363 | 366-559 |
| SmCWIN2 | HU02G03468.1 | 细胞壁Cell wall | 1-19 | 48-342 | 345-538 |
| SmCWIN3 | HU07G01984.1 | 细胞壁Cell wall | 1-27 | 46-366 | 369-562 |
| SmCWIN4 | HU07G01985.1 | 细胞质Cytoplasm | - | 10-327 | 330-523 |
| SmCWIN5 | HU07G01986.1 | 细胞壁Cell wall | 1-34 | 61-378 | 381-575 |
| SmCWIN6 | HU09G00322.1 | 细胞壁Cell wall | 1-31 | 61-380 | 383-576 |
| SmCWIN7 | HU09G01009.1 | 细胞质Cytoplasm | - | 7-104 | 107-287 |
Table 2 Sequence prediction of SmCWIN gene family from red pitaya
名称 Name | 基因ID Gene ID | 定位 Location | 位置 Position (aa) | ||
|---|---|---|---|---|---|
信号肽 Signal peptide | GH32N结构域 GH32N domain | GH32C结构域 GH32C domain | |||
| SmCWIN1 | HU02G02988.1 | 细胞壁Cell wall | 1-19 | 48-363 | 366-559 |
| SmCWIN2 | HU02G03468.1 | 细胞壁Cell wall | 1-19 | 48-342 | 345-538 |
| SmCWIN3 | HU07G01984.1 | 细胞壁Cell wall | 1-27 | 46-366 | 369-562 |
| SmCWIN4 | HU07G01985.1 | 细胞质Cytoplasm | - | 10-327 | 330-523 |
| SmCWIN5 | HU07G01986.1 | 细胞壁Cell wall | 1-34 | 61-378 | 381-575 |
| SmCWIN6 | HU09G00322.1 | 细胞壁Cell wall | 1-31 | 61-380 | 383-576 |
| SmCWIN7 | HU09G01009.1 | 细胞质Cytoplasm | - | 7-104 | 107-287 |
蛋白 Protein | 位点/结构域位置 Site or domain position (aa) | |||||||
|---|---|---|---|---|---|---|---|---|
| 63 | 65-69 | 90 | 125 | 191-193 | 245-249 | 282 | 285 | |
| AtcwINV1 | W | NDPNG | W | W | RDP | WECPD | D | K |
| SmCWIN1 | W | N - - - G | W | Y | RDP | WECPD | V | - |
| SmCWIN2 | W | N - - - G | W | Y | RDP | WECPD | V | - |
| SmCWIN3 | W | NDPNG | W | W | RDP | WECPD | F | S |
| SmCWIN4 | W | N - - - G | W | W | RDP | WECPD | F | L |
| SmCWIN5 | W | N - - - A | W | W | RDP | WECPD | D | K |
| SmCWIN6 | W | NDPNG | W | W | RDP | WECPD | D | K |
| SmCWIN7 | - | - - - - - | - | - | - - - | - - - - - | M | D |
Table 3 Conserved site and domain analysis of amino acid sequences of SmCWINs
蛋白 Protein | 位点/结构域位置 Site or domain position (aa) | |||||||
|---|---|---|---|---|---|---|---|---|
| 63 | 65-69 | 90 | 125 | 191-193 | 245-249 | 282 | 285 | |
| AtcwINV1 | W | NDPNG | W | W | RDP | WECPD | D | K |
| SmCWIN1 | W | N - - - G | W | Y | RDP | WECPD | V | - |
| SmCWIN2 | W | N - - - G | W | Y | RDP | WECPD | V | - |
| SmCWIN3 | W | NDPNG | W | W | RDP | WECPD | F | S |
| SmCWIN4 | W | N - - - G | W | W | RDP | WECPD | F | L |
| SmCWIN5 | W | N - - - A | W | W | RDP | WECPD | D | K |
| SmCWIN6 | W | NDPNG | W | W | RDP | WECPD | D | K |
| SmCWIN7 | - | - - - - - | - | - | - - - | - - - - - | M | D |
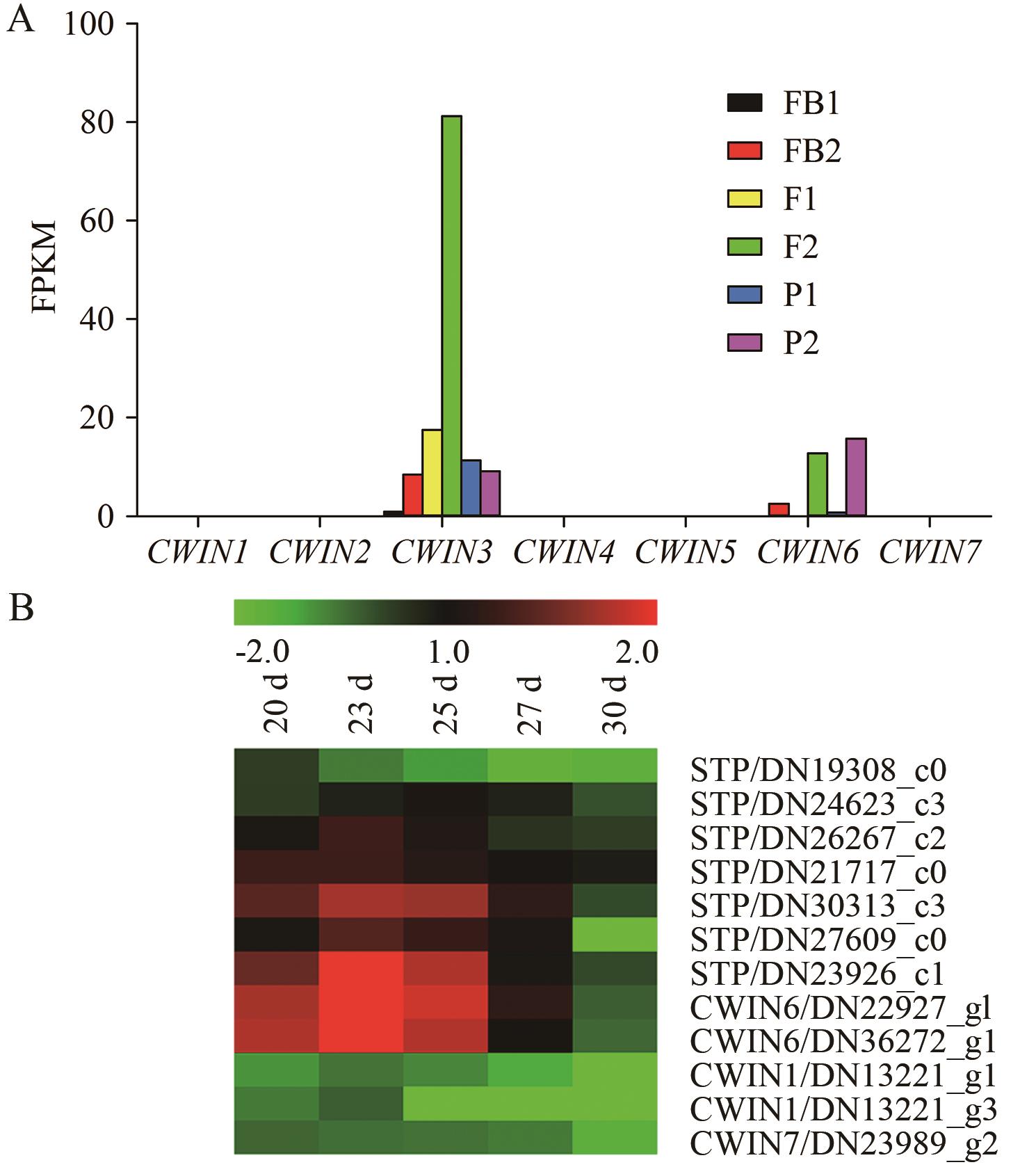
Fig. 3 Expression patterns of CWINs and STPs in red pitayaA: Expression patterns of CWINs during flower bud differentiation, flowering and fruit peel development. FB1-FB2: Two stages during flower bud differentiation. F1-F2: Two stages during flowering. P1-P2: Two stages during fruit peel development. B: Expression patterns of CWINs and STPsduring fruit pulp development. FPKM values of A and B came from the pitaya genome database (http://pitayagenomic.com/) and red pitaya fruit transcriptome sequencing [23], respectively
| [1] | Khan D, Harris A, Zaman QU, et al. The evolutionary history and distribution of Cactus germplasm resources, as well as potential domestication under a changing climate [J]. J Syst Evol, 2024, 62(5): 858-875. |
| [2] | Shah K, Chen JY, Chen JX, et al. Pitaya nutrition, biology, and biotechnology: a review [J]. Int J Mol Sci, 2023, 24(18): 13986. |
| [3] | Li J, Foster R, Ma S, et al. Identification of transcription factors controlling cell wall invertase gene expression for reproductive development via bioinformatic and transgenic analyses [J]. Plant J, 2021, 106(4): 1058-1074. |
| [4] | Wan HJ, Wu LM, Yang YJ, et al. Evolution of sucrose metabolism: the dichotomy of invertases and beyond [J]. Trends Plant Sci, 2018, 23(2): 163-177. |
| [5] | 王海波, 辛胡, 刘潮, 等. 小桐子转化酶基因家族的生物信息学分析及表达特性 [J]. 生物技术通报, 2017, 33(9): 172-178. |
| Wang HB, Xin H, Liu C, et al. Genome-wide identification and expression characteristics of invertase gene family in Jatropha curcas [J]. Biotechnol Bull, 2017, 33(9): 172-178. | |
| [6] | Wang L, Ruan YL. New insights into roles of cell wall invertase in early seed development revealed by comprehensive spatial and temporal expression patterns of GhCWIN1 in cotton [J]. Plant Physiol, 2012, 160(2): 777-787. |
| [7] | Zhang JQ, Wu ZC, Hu FC, et al. Aberrant seed development in Litchi chinensis is associated with the impaired expression of cell wall invertase genes [J]. Hortic Res, 2018, 5: 39. |
| [8] | Seitz J, Reimann TM, Fritz C, et al. How pollen tubes fight for food: the impact of sucrose carriers and invertases of Arabidopsis thaliana on pollen development and pollen tube growth [J]. Front Plant Sci, 2023, 14: 1063765. |
| [9] | Li B, Liu H, Zhang Y, et al. Constitutive expression of cell wall invertase genes increases grain yield and starch content in maize [J]. Plant Biotechnol J, 2013, 11(9): 1080-1091. |
| [10] | Yan W, Wu XY, Li YN, et al. Cell wall invertase 3 affects cassava productivity via regulating sugar allocation from source to sink [J]. Front Plant Sci, 2019, 10: 541. |
| [11] | Liu H, Yao XH, Fan JW, et al. Cell wall invertase 3 plays critical roles in providing sugars during pollination and fertilization in cucumber [J]. Plant Physiol, 2024, 195(2): 1293-1311. |
| [12] | Ren ZM, Zhang D, Jiao C, et al. Comparative transcriptome and metabolome analyses identified the mode of sucrose degradation as a metabolic marker for early vegetative propagation in bulbs of Lycoris [J]. Plant J, 2022, 112(1): 115-134. |
| [13] | Palmer WM, Ru L, Jin Y, et al. Tomato ovary-to-fruit transition is characterized by a spatial shift of mRNAs for cell wall invertase and its inhibitor with the encoded proteins localized to sieve elements [J]. Mol Plant, 2015, 8(2): 315-328. |
| [14] | Kawaguchi K, Takei-Hoshi R, Yoshikawa I, et al. Functional disruption of cell wall invertase inhibitor by genome editing increases sugar content of tomato fruit without decrease fruit weight [J]. Sci Rep, 2021, 11(1): 21534. |
| [15] | Lou HC, Li SJ, Shi ZH, et al. Engineering source-sink relations by prime editing confers heat-stress resilience in tomato and rice [J]. Cell, 2025, 188(2): 530-549.e20. |
| [16] | Yuan HZ, Pang FH, Cai WJ, et al. Genome-wide analysis of the invertase genes in strawberry (Fragaria × Ananassa) [J]. J Integr Agric, 2021, 20(10): 2652-2665. |
| [17] | Gao YY, Yao YL, Chen X, et al. Metabolomic and transcriptomic analyses reveal the mechanism of sweet-acidic taste formation during pineapple fruit development [J]. Front Plant Sci, 2022, 13: 971506. |
| [18] | Hua QZ, Chen CB, Tel Zur N, et al. Metabolomic characterization of pitaya fruit from three red-skinned cultivars with different pulp colors [J]. Plant Physiol Biochem, 2018, 126: 117-125. |
| [19] | Wei W, Cheng MN, Ba LJ, et al. Pitaya HpWRKY3 is associated with fruit sugar accumulation by transcriptionally modulating sucrose metabolic genes HpINV2 and HpSuSy1 [J]. Int J Mol Sci, 2019, 20(8): 1890. |
| [20] | Zhang ZK, Xing YM, Ramakrishnan M, et al. Transcriptomics-based identification and characterization of genes related to sugar metabolism in ‘Hongshuijing’ pitaya [J]. Hortic Plant J, 2022, 8(4): 450-460. |
| [21] | 郑乾明, 晏霜, 王红林, 等. 红肉火龙果液泡转化酶基因HpVIN1的克隆、表达和酶活性鉴定 [J]. 热带作物学报, 2024, 45(12): 2524-2533. |
| Zheng QM, Yan S, Wang HL, et al. Cloning, expression and enzyme activity identification of the vacuolar invertase gene HpVIN1 from red pitaya [J]. Chin J Trop Crops, 2024, 45(12): 2524-2533. | |
| [22] | 杨器, 李广泽, 杨成坤, 等. 火龙果酸性转化酶基因家族HuAI的鉴定及生物信息学分析 [J/OL]. 分子植物育种, 2022. . |
| Yang Q, Li GZ, Yang CK, et al. Identification and bioinformatics analysis of acid invertase gene family in pitaya [J/OL]. Mol Plant Breed, 2022. . | |
| [23] | Zheng QM, Wang XK, Qi Y, et al. Selection and validation of reference genes for qRT-PCR analysis during fruit ripening of red pitaya (Hylocereus polyrhizus) [J]. FEBS Open Bio, 2021, 11(11): 3142-3152. |
| [24] | Wu TM, Lin KC, Liau WS, et al. A set of GFP-based organelle marker lines combined with DsRed-based gateway vectors for subcellular localization study in rice (Oryza sativa L.) [J]. Plant Mol Biol, 2016, 90(1/2): 107-115. |
| [25] | De Coninck B, Le Roy K, Francis I, et al. Arabidopsis AtcwINV3 and 6 are not invertases but are fructan exohydrolases (FEHs) with different substrate specificities [J]. Plant Cell Environ, 2005, 28(4): 432-443. |
| [26] | Roitsch T, González MC. Function and regulation of plant invertases: sweet sensations [J]. Trends Plant Sci, 2004, 9(12): 606-613. |
| [27] | Chen TH, Huang YC, Yang CS, et al. Insights into the catalytic properties of bamboo vacuolar invertase through mutational analysis of active site residues [J]. Phytochemistry, 2009, 70(1): 25-31. |
| [28] | Shen LB, Qin YL, Qi ZQ, et al. Genome-wide analysis, expression profile, and characterization of the acid invertase gene family in pepper [J]. Int J Mol Sci, 2018, 20(1): 15. |
| [29] | Li YM, Forney C, Bondada B, et al. The molecular regulation of carbon sink strength in grapevine (Vitis vinifera L.) [J]. Front Plant Sci, 2021, 11: 606918. |
| [30] | Le Roy K, Lammens W, Verhaest M, et al. Unraveling the difference between invertases and fructan exohydrolases: a single amino acid (Asp-239) substitution transforms Arabidopsis cell wall invertase1 into a fructan 1-exohydrolase [J]. Plant Physiol, 2007, 145(3): 616-625. |
| [31] | Chen L, Zheng FH, Feng ZL, et al. A vacuolar invertase CsVI2 regulates sucrose metabolism and increases drought tolerance in Cucumis sativus L [J]. Int J Mol Sci, 2021, 23(1): 176. |
| [32] | Zhang K, Wu ZD, Wu XL, et al. Regulatory and functional divergence among members of Ibβfruct2, a sweet potato vacuolar invertase gene controlling starch and glucose content [J]. Front Plant Sci, 2023, 14: 1192417. |
| [33] | Huang DM, Wu B, Chen G, et al. Genome-wide analysis of the passion fruit invertase gene family reveals involvement of PeCWINV5 in hexose accumulation [J]. BMC Plant Biol, 2024, 24(1): 836. |
| [34] | Li MJ, Feng FJ, Cheng LL. Expression patterns of genes involved in sugar metabolism and accumulation during apple fruit development [J]. PLoS One, 2012, 7(3): e33055. |
| [35] | McCurdy DW, Dibley S, Cahyanegara R, et al. Functional characterization and RNAi-mediated suppression reveals roles for hexose transporters in sugar accumulation by tomato fruit [J]. Mol Plant, 2010, 3(6): 1049-1063. |
| [36] | Ru L, He Y, Zhu ZJ, et al. Integrating sugar metabolism with transport: elevation of endogenous cell wall invertase activity up-regulates SlHT2 and SlSWEET12c expression for early fruit development in tomato [J]. Front Genet, 2020, 11: 592596. |
| [37] | Wang ZY, Wei XY, Yang JJ, et al. Heterologous expression of the apple hexose transporter MdHT2.2 altered sugar concentration with increasing cell wall invertase activity in tomato fruit [J]. Plant Biotechnol J, 2020, 18(2): 540-552. |
| [38] | 郑乾明, 晏霜, 解璞, 等. 红肉火龙果糖转运蛋白HpSWEET4b基因克隆、表达和转运活性鉴定 [J]. 西南农业学报, 2024, 37(12): 2739-2748. |
| Zheng QM, Yan S, Xie P, et al. Cloning, expression analysis and transport activity assay of sugar transporter gene HpSWEET4b in red pitaya (Hylocereus polyrhizus) [J]. Southwest China J Agric Sci, 2024, 37(12): 2739-2748. |
| [1] | WU Shuang, LU Rui-lin, FENG Cheng-tian, YUAN Kun, WANG Zhen-hui, LIU Jin-ping, LIU Hui. Expression of HbTRXh5 Gene of Hevea brasiliensis in Yeast and Analysis on Its Resistance to Stress [J]. Biotechnology Bulletin, 2024, 40(12): 136-144. |
| [2] | GONG Yuan-yong, ZHAO Li-hua, YAN Fei, GUO Shu-qiao, SHU Hong-mei, NI Wan-chao. Construction and Transformation of Borago officinalis BoD6D Gene Vector [J]. Biotechnology Bulletin, 2021, 37(3): 227-232. |
| [3] | LIU Hui, DENG Zhi, YANG Hong, DAI Long-jun, LI De-jun. Expression and Stress Tolerance Analysis of HbMC2 Gene from Hevea brasliensis in Yeast [J]. Biotechnology Bulletin, 2018, 34(9): 202-208. |
| [4] | Wang Haibo, Zou Zhurong, Gong Ming. Gene Cloning, Bioinformatic Analysis and Preliminary Functional Characterization of Gene Encoding Galactinol Synthase 3 in Jatropha curcas [J]. Biotechnology Bulletin, 2015, 31(7): 91-100. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||