








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (11): 177-189.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0392
XU Yuan-zhi1,2( ), HU Shan2, DAI Si-ze2, YOU Shuai1, ZHENG Ming-ming1,2(
), HU Shan2, DAI Si-ze2, YOU Shuai1, ZHENG Ming-ming1,2( ), SHAN Kai3
), SHAN Kai3
Received:2025-04-17
Online:2025-11-26
Published:2025-12-09
Contact:
ZHENG Ming-ming
E-mail:2292385667@qq.com;zhengmingming@caas.cn
XU Yuan-zhi, HU Shan, DAI Si-ze, YOU Shuai, ZHENG Ming-ming, SHAN Kai. Discovery and Evaluation of High-activity Lipase and Its Application in Diacylglycerol Synthesis[J]. Biotechnology Bulletin, 2025, 41(11): 177-189.
引物名称 Primer name | 引物序列 Sequence of primer(5′-3′) | 功能 Function |
|---|---|---|
| 27-F | AGAGTTTGATCCTGGCTCAG | 扩增16S rDNA序列用于菌种鉴定 |
| 1492-R | TACGGCTACCTTGTTACGACTT | |
| F | CGTGCTTCACTCCGCATT | 扩增脂肪酶前段序列bclipF |
| R | GACCGGGTCTTCCGCA | |
| 12C-F | TTCGTCAATGTATTCGGCA | 扩增脂肪酶后段序列bclipR |
| 11-R | GAGCGCATCGAGATACGC | |
| KZ-F | CAAATGGGTCGCGGATCC | 扩增完整脂肪酶序列bclip12c |
| KZ-R | TCGAGTGCGGCCGC |
Table 1 Main primers used in PCR
引物名称 Primer name | 引物序列 Sequence of primer(5′-3′) | 功能 Function |
|---|---|---|
| 27-F | AGAGTTTGATCCTGGCTCAG | 扩增16S rDNA序列用于菌种鉴定 |
| 1492-R | TACGGCTACCTTGTTACGACTT | |
| F | CGTGCTTCACTCCGCATT | 扩增脂肪酶前段序列bclipF |
| R | GACCGGGTCTTCCGCA | |
| 12C-F | TTCGTCAATGTATTCGGCA | 扩增脂肪酶后段序列bclipR |
| 11-R | GAGCGCATCGAGATACGC | |
| KZ-F | CAAATGGGTCGCGGATCC | 扩增完整脂肪酶序列bclip12c |
| KZ-R | TCGAGTGCGGCCGC |
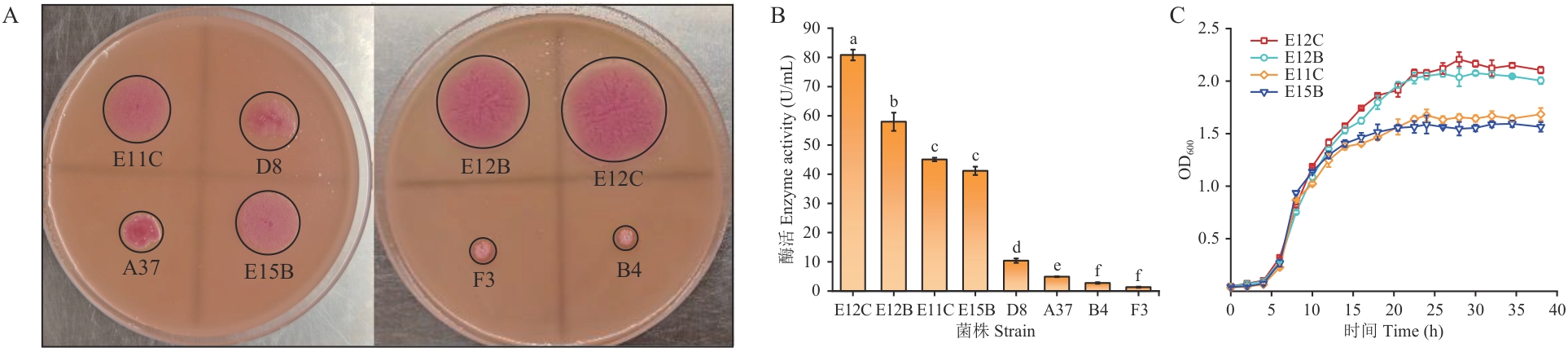
Fig. 1 Excavation and growth curve of lipase-producing strainsA: Discoloration circle of lipase-producing strains on the plate. B: Lipase activity of each strain. C: Strain growth curve. Different letters indicate significant differences (P≤0.05), the same below

Fig. 2 Identification of strain E12CA: Colony morphology of E12C strain on LB plate. B: Scanning electron microscope at 400 nm. C: Phylogenetic tree of16S rDNA for strain E12C
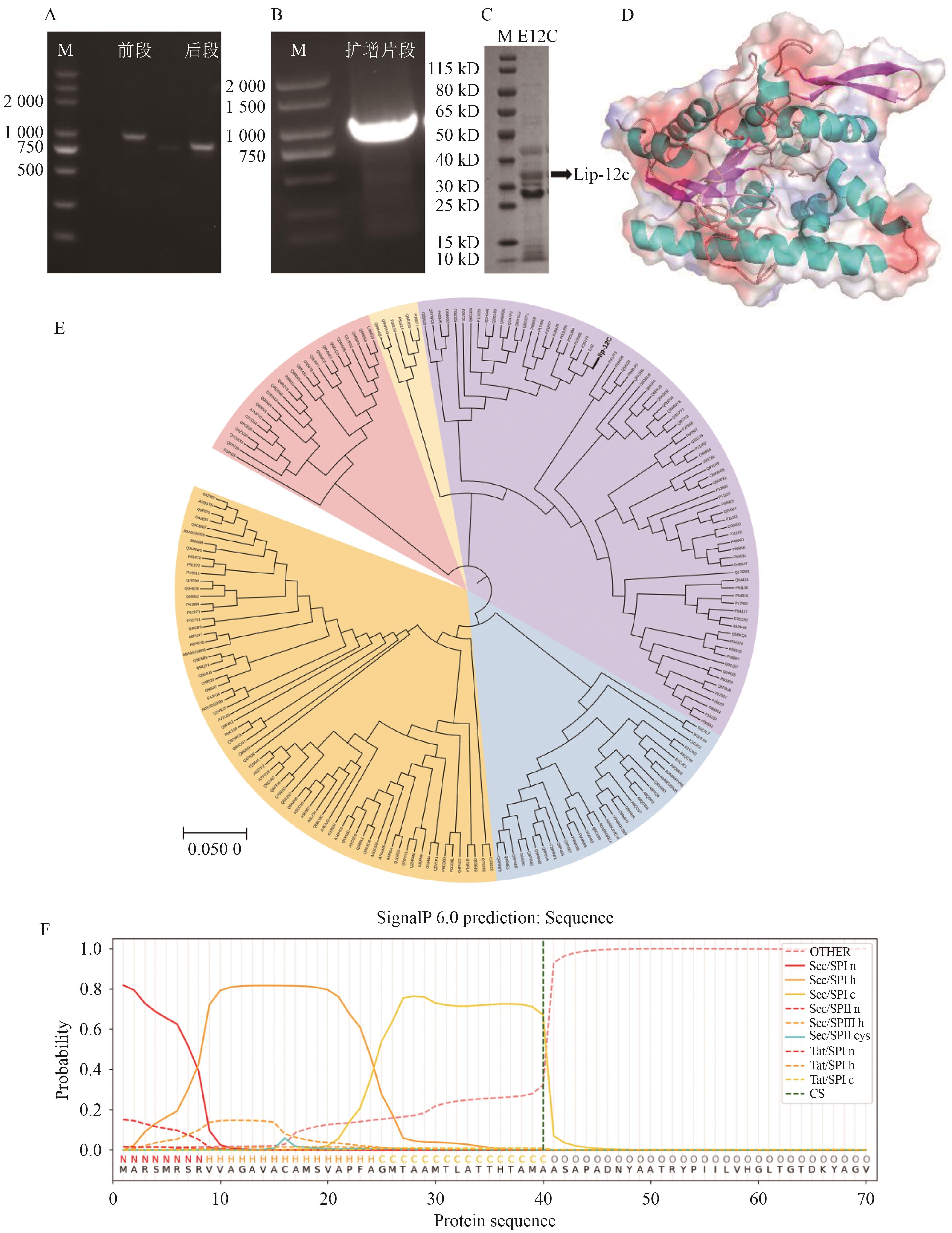
Fig. 3 Identification of lipase Lip-12c geneA: Lipase gene sequence bclipF and bclipR. B: Mature lipase gene sequence bclip12c. C: Analysis of SDS-PAGE results of lipase Lip-12c. D: Predicted three-dimensional structure of lipase Lip-12c. E: Phylogenetic tree of lipase Lip-12c. F: Signal peptide prediction results
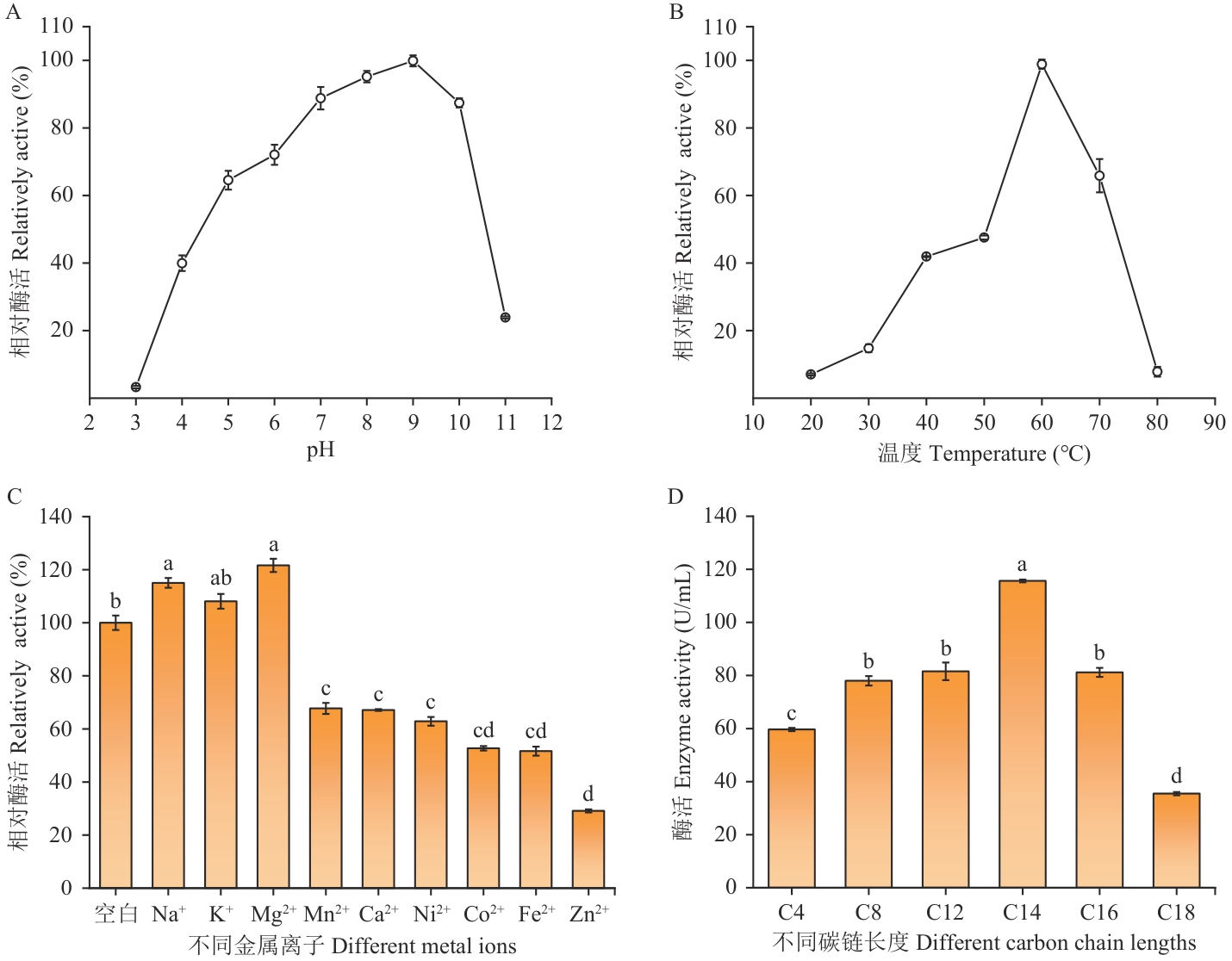
Fig. 4 Analysis of enzymatic propertiesA: Effect of pH on enzyme activity. B: Effect of temperature on enzyme activity. C: Effect of metal ions on enzyme activity. D: Substrate chain length specificity
有机溶剂 Organic solvent | 油水分配系数 Log P | 相对酶活 Relative activity (%) | 有机溶剂 Organic solvent | 油水分配系数 Log P | 相对酶活 Relative activity (%) |
|---|---|---|---|---|---|
| 无有机溶剂 | - | 100 ± 1.44ab | 乙酸 | -0.17 | 70.62 ± 1.08g |
| 正己烷 | 3.5 | 85.37 ± 1.77de | 丙酮 | -0.24 | 77.44 ± 2.71f |
| 环己烷 | 3.44 | 88.50 ± 1.03d | 乙醇 | -0.31 | 91.69 ± 1.48c |
| 二氯甲烷 | 1.25 | 77.01 ± 0.71f | 乙腈 | -0.33 | 90.66 ± 2.97cd |
| 乙醚 | 0.85 | 103.68 ± 1.29a | 甲醇 | -0.77 | 97.23 ± 0.76bc |
| 乙酸乙酯 | 0.68 | 81.44 ± 0.64e | 丙三醇 | -1.76 | 87.42 ± 0.74d |
| 异丙醇 | 0.05 | 79.94 ± 1.16ef |
Table 2 Effect of organic solvents on the activity of lipase lip-12c
有机溶剂 Organic solvent | 油水分配系数 Log P | 相对酶活 Relative activity (%) | 有机溶剂 Organic solvent | 油水分配系数 Log P | 相对酶活 Relative activity (%) |
|---|---|---|---|---|---|
| 无有机溶剂 | - | 100 ± 1.44ab | 乙酸 | -0.17 | 70.62 ± 1.08g |
| 正己烷 | 3.5 | 85.37 ± 1.77de | 丙酮 | -0.24 | 77.44 ± 2.71f |
| 环己烷 | 3.44 | 88.50 ± 1.03d | 乙醇 | -0.31 | 91.69 ± 1.48c |
| 二氯甲烷 | 1.25 | 77.01 ± 0.71f | 乙腈 | -0.33 | 90.66 ± 2.97cd |
| 乙醚 | 0.85 | 103.68 ± 1.29a | 甲醇 | -0.77 | 97.23 ± 0.76bc |
| 乙酸乙酯 | 0.68 | 81.44 ± 0.64e | 丙三醇 | -1.76 | 87.42 ± 0.74d |
| 异丙醇 | 0.05 | 79.94 ± 1.16ef |
脂肪酶 Lipase | 蛋白浓度 Protein concentration (mg/mL) | 比活力 Specific activity (U/g protein) | 酶来源 Enzyme source |
|---|---|---|---|
| Lip-12c | 1.57 ± 0.04b | 39 254.5 ± 271.3a | 本研究 |
| Candida rugosa lipase AY 400SD | 2.49 ± 0.12a | 6 171.1 ± 507.3b | 商品酶 |
| C. rugosa lipase CRL | 1.21 ± 0.18b | 3 376.6 ± 223.3c | |
| C. rugosa lipase AY SD | 0.27 ± 0.05c | 298.0 ± 31.3d | |
| C. rugosa lipase AYS | 0.26 ± 0.05c | 220.1 ± 15.5d | |
| Geobacillus thermocatenulatus KCTC | - | 22 700 | [ |
| Aeromonas caviae LipT51 | - | 34 200 | [ |
Table 3 Comparison of specific activity of different lipases
脂肪酶 Lipase | 蛋白浓度 Protein concentration (mg/mL) | 比活力 Specific activity (U/g protein) | 酶来源 Enzyme source |
|---|---|---|---|
| Lip-12c | 1.57 ± 0.04b | 39 254.5 ± 271.3a | 本研究 |
| Candida rugosa lipase AY 400SD | 2.49 ± 0.12a | 6 171.1 ± 507.3b | 商品酶 |
| C. rugosa lipase CRL | 1.21 ± 0.18b | 3 376.6 ± 223.3c | |
| C. rugosa lipase AY SD | 0.27 ± 0.05c | 298.0 ± 31.3d | |
| C. rugosa lipase AYS | 0.26 ± 0.05c | 220.1 ± 15.5d | |
| Geobacillus thermocatenulatus KCTC | - | 22 700 | [ |
| Aeromonas caviae LipT51 | - | 34 200 | [ |
| [15] | Chandra P, Enespa, Singh R, et al. Microbial lipases and their industrial applications: a comprehensive review [J]. Microb Cell Fact, 2020, 19(1): 169. |
| [16] | 张静, 贾佳, 董帅豪, 等. 国产毕赤酵母菌脂肪酶催化酯交换制备零反式、低饱和人造奶油基料油 [J]. 中国油脂, 2024, 49(8): 120-126. |
| Zhang J, Jia J, Dong SH, et al. Preparation of zero trans, low saturated margarine base oil by transesterification catalyzed by domestic Pichia pastoris lipase [J]. China Oils and Fats, 2024, 49(8): 120-126. | |
| [17] | 徐碧林, 朱庆. 微生物脂肪酶稳定性研究进展 [J]. 微生物学通报, 2020, 47(6): 1958-1972. |
| Xu BL, Zhu Q. Recent advances in stability studies of microbial lipase [J]. Microbiol China, 2020, 47(6): 1958-1972. | |
| [18] | Mahadik ND, Puntambekar US, Bastawde KB, et al. Production of acidic lipase by Aspergillus niger in solid state fermentation [J]. Process Biochem, 2002, 38(5): 715-721. |
| [19] | 布坎南 吉本斯. 伯杰细菌鉴定手册: 第八版 [M]. 8版. 北京: 科学出版社, 1984. |
| Buchanan G. Bergey’s manual of determinative bacteriology [M]. 8th ed. Beijing: Science Press, 1984. | |
| [20] | 东秀珠, 蔡妙英. 常见细菌系统鉴定手册 [M]. 北京: 科学出版社, 2001. |
| Dong XZ, Cai MY. Handbook of identification of common bacterial systems [M]. Beijing: Science Press, 2001. | |
| [21] | 贾彬. 洋葱伯克霍尔德菌的筛选、鉴定及其脂肪酶基因的高效表达 [D]. 武汉: 华中科技大学, 2010. |
| Jia B. Screening and identification of Burkholderia cepacia and its high-level expression of lipase gene [D]. Wuhan: Huazhong University of Science and Technology, 2010. | |
| [22] | Xu LJ, Zhang Y, Zivkovic V, et al. Deacidification of high-acid rice bran oil by the tandem continuous-flow enzymatic reactors [J]. Food Chem, 2022, 393: 133440. |
| [23] | Mezzetti A, Schrag JD, Cheong CS, et al. Mirror-image packing in enantiomer discrimination molecular basis for the enantioselectivity of B.cepacia lipase toward 2-methyl-3-phenyl-1-propanol [J]. Chem Biol, 2005, 12(4): 427-437. |
| [24] | Gurkok S, Ozdal M. Purification and characterization of a novel extracellular, alkaline, thermoactive, and detergent-compatible lipase from Aeromonas caviae LipT51 for application in detergent industry [J]. Protein Expr Purif, 2021, 180: 105819. |
| [25] | Jo E, Kim J, Lee A, et al. Identification and characterization of a novel thermostable GDSL-type lipase from Geobacillus thermocatenulatus [J]. J Microbiol Biotechnol, 2021, 31(3): 483-491. |
| [26] | 李桥妹, 王宗德, 谢礼文, 等. 酶法制备富含中碳链甘油二酯的山苍子核仁油工艺研究 [J]. 中国粮油学报, 2021, 36(8): 54-59. |
| Li QM, Wang ZD, Xie LW, et al. Preparation of litseacubeba kernel oil rich in medium-chain diacylglycerol byenzymaticmethod [J]. J Chin Cereals Oils Assoc, 2021, 36(8): 54-59. | |
| [27] | Liu R, Zhang Y, Xu YZ, et al. Molecular docking simulation reveals the lipase-substrate binding mechanism in the enzymatic synthesis of diacylglycerol-enriched vegetable oils [J]. Food Chem, 2025, 474: 143236. |
| [28] | Sánchez DA, Tonetto GM, Ferreira ML. Burkholderia cepacia lipase: a versatile catalyst in synthesis reactions [J]. Biotechnol Bioeng, 2018, 115(1): 6-24. |
| [29] | 秦蓁天, 王小兵, 汪晓丽, 等. 餐厨垃圾高效油脂降解菌的筛选及其降解条件研究 [J]. 扬州大学学报: 农业与生命科学版, 2023, 44(5): 118-126, 134. |
| Qin ZT, Wang XB, Wang XL, et al. Screening of high-efficiency oil-degrading bacteria from kitchen waste and study on its degradation conditions [J]. Journal of Yangzhou University: Agricultural and Life Science Edition, 2023, 44(5): 118-126, 134. | |
| [30] | Tavares M, Kozak M, Balola A, et al. Burkholderia cepacia complex bacteria: a feared contamination risk in water-based pharmaceutical products [J]. Clin Microbiol Rev, 2020, 33(3): e00139-19. |
| [31] | 李仰龙, 魏书蒙, 陈详腾, 等. 洋葱伯克霍尔德菌菌株JLS 17对Cd2+胁迫的生长及代谢响应机制 [J]. 林业科学, 2024, 60(8): 152-163. |
| [1] | Xu LL, Li JX, Zhang HJ, et al. Biological modification and industrial applications of microbial lipases: a general review [J]. Int J Biol Macromol, 2025, 302: 140486. |
| [2] | 陈善辉, 于爱霞, 王东, 等. 甘油二酯安全性、代谢及减肥降脂功能研究进展 [J]. 食品工业, 2024, 45(11): 211-217. |
| Chen SH, Yu AX, Wang D, et al. Research progress on safety, metabolism, weight loss and lipid-lowering function of diglycerides [J]. The Food Industry, 2024, 45(11): 211-217. | |
| [3] | Zeng Q, Sun MM, Xie X, et al. Lipase-entrapped colloidosomes with tunable positioning at the oil-water interface for Pickering emulsion-enhanced biocatalysis [J]. ACS Appl Mater Interfaces, 2022, 14(49): 54781-54789. |
| [4] | Akram F, Mir AS, Haq IU, et al. An appraisal on prominent industrial and biotechnological applications of bacterial lipases [J]. Mol Biotechnol, 2023, 65(4): 521-543. |
| [5] | Yan QF, Li ZY, Sun RJ, et al. Promoted expression of a lipase for its application in EPA/DHA enrichment and mechanistic insights into its substrate specificity [J]. Int J Biol Macromol, 2025, 296: 139628. |
| [6] | Wang J, Liu X, Wang XD, et al. Selective synthesis of human milk fat-style structured triglycerides from microalgal oil in a microfluidic reactor packed with immobilized lipase [J]. Bioresour Technol, 2016, 220: 132-141. |
| [7] | Zhang TT, Zhang Y, Deng CY, et al. Green and efficient synthesis of highly liposoluble and antioxidant L-ascorbyl esters by immobilized lipases [J]. J Clean Prod, 2022, 379: 134772. |
| [8] | Zhang SY, Hyatt JR, Akoh CC. Lipase-catalyzed one-step regioselective synthesis of 1, 2-dioctanoylgalloylglycerol in a solvent-free system: Optimization of reaction conditions and structural elucidation [J]. Food Chem, 2022, 382: 132302. |
| [9] | Zhang YF, Lai YD, Zheng MM. Ultrasound-assisted intensification of Pickering interfacial biocatalysis preparation of vitamin A aliphatic esters [J]. Ultrason Sonochem, 2024, 107: 106929. |
| [10] | Li DY, Chen XY, Chen ZC, et al. Directed evolution of lipase A from Bacillus subtilis for the preparation of enantiocomplementary sec-alcohols [J]. Green Synth Catal, 2021, 2(3): 290-294. |
| [11] | Alabdalall AH, Al-Anazi NA, Aldakheel LA, et al. Application and characterization of crude fungal lipases used to degrade fat and oil wastes [J]. Sci Rep, 2021, 11(1): 19670. |
| [12] | Yang XX, Zhang Y, Pang HM, et al. Codisplay of Rhizopus oryzae and Candida rugosa lipases for biodiesel production [J]. Catalysts, 2021, 11(4): 421. |
| [13] | Zulaika A, Rahman H, Ningrum SS, et al. Exploring microbial lipases: Screening and identification for biocatalyst potential in bioethanol synthesis from glycerol-based biodiesel waste [J]. Results Eng, 2024, 23: 102427. |
| [14] | Ismail AR, Kashtoh H, Baek KH. Temperature-resistant and solvent-tolerant lipases as industrial biocatalysts: Biotechnological approaches and applications [J]. Int J Biol Macromol, 2021, 187: 127-142. |
| [31] | Li YL, Wei SM, Chen XT, et al. Growth and metabolic response mechanism of Burkholderia cepacia strain JLS17 to Cd2+ stress [J]. Scientia Silvae Sinicae, 2024, 60(8): 152-163. |
| [32] | Chen G, Zhang QP, Chen HT, et al. In situ and real-time insight into Rhizopus chinensis lipase under high pressure and temperature: Conformational traits and biobehavioural analysis [J]. Int J Biol Macromol, 2020, 154: 1314-1323. |
| [33] | Abd Razak NN, Cognet P, Pérès Y, et al. Kinetics and hydrodynamics of Candida antartica lipase-catalyzed synthesis of glycerol dioleate (GDO) in a continuous flow packed-bed millireactor [J]. J Clean Prod, 2022, 373: 133816. |
| [34] | Rasmussen HØ, Wollenberg DTW, Wang HB, et al. The changing face of SDS denaturation: complexes of Thermomyces lanuginosus lipase with SDS at pH 4.0, 6.0 and 8.0 [J]. J Colloid Interface Sci, 2022, 614: 214-232. |
| [35] | Karaman F, Incekara U, Arslan NP, et al. A new enzyme for biodiesel production and food applications: lipase of Bacillus megateriumF25 isolated from an aquatic insect Rhantus suturalis [J]. GCB Bioenergy, 2024, 16(12): e70009. |
| [36] | Karabulut I, Durmaz G, Hayaloglu AA. Fatty acid selectivity of lipases during acidolysis reaction between triolein and saturated fatty acids varying from caproic to behenic acids [J]. J Agric Food Chem, 2009, 57(16): 7584-7590. |
| [37] | Cong YX, Liu YW, Yuan MY, et al. Efficient preparation of diglycerides from rapeseed oil sediment using phospholipase C in a solvent-free phospholipid hydrolysis system [J]. Food Biosci, 2024, 62: 105175. |
| [38] | Liu YS, Yang ZH, Zhou XM, et al. Diacylglycerol kinases and its role in lipid metabolism and related diseases [J]. Int J Mol Sci, 2024, 25(23): 13207. |
| [1] | SU Xiu-min, HAN Wen-qing, WANG Jiao, LI Peng, WANG Qiu-lan, LI Wan-xing, CAO Jin-jun. Isolation, Identification, Biological Characteristics and Biocontrol Effects of Trichoderma harzianum M408 against Tomato Early Blight [J]. Biotechnology Bulletin, 2025, 41(9): 277-288. |
| [2] | LIAN Shao-jie, TANG Sheng-shuo, KANG Chuan-li, LIU Lei, ZHENG De-qiang, DU Shuai, TANG Li-wei, ZHANG Mei-xia, LIU Qiang. Isolation, Identification, Optimization of Fermentation Conditions of High-yield Tremella fuciformis Polysaccharides Enzyme-producing Strain and Its Enzyme Characteristics Analysis [J]. Biotechnology Bulletin, 2025, 41(9): 302-313. |
| [3] | ZHANG Ru, LI Yi-ming, ZHANG Tong-xi, SUN Zhan-bin, REN Qing, PAN Han-xu. Isolation and Identification of a High-yielding Magnolol and Honokiol Strain from Magnolia officinalis and Optimization of the “Sweating” Process [J]. Biotechnology Bulletin, 2025, 41(8): 322-334. |
| [4] | XIANG Bo-ka, ZHOU Zuan-zuan, FENG Jia-hui, XIA Chen, LI Qi, CHEN Chun. Isolation and Identification of a Fungus from Moldy Tobacco Leaf and Study on Its Mold-causing Factors [J]. Biotechnology Bulletin, 2025, 41(2): 321-330. |
| [5] | ZHANG Ya-ya, LI Pan-pan, GAO Hui-hui, JIA Chen-bo, XU Chun-yan. Exploring on the Pathogenesis of Root Rot of Lycium barbarum cv. ‘Ningqi-5' Based on the Rhizoplane Fungal Community and Pathogens Identification [J]. Biotechnology Bulletin, 2024, 40(9): 238-248. |
| [6] | CUI Yuan-yuan, WANG Zhao-yi, BAI Shuang-yu, REN Yu-zhao, DOU Fei-fei, LIU Cai-xia, LIU Feng-lou, WANG Zhang-jun, LI Qing-feng. Genome-wide Identification of Non-specific Phospholipase C Gene Family in Hordeum vulgare L. and Stress Expression Analysis at Seedling Stage [J]. Biotechnology Bulletin, 2024, 40(8): 74-82. |
| [7] | WANG Fang, YU Lu, QI Ze-zheng, ZHOU Chang-jun, YU Ji-dong. Screening and Biocontrol Effect of Antagonistic Bacteria against Soybean Root Rot [J]. Biotechnology Bulletin, 2024, 40(7): 216-225. |
| [8] | YIN Liang, WANG Dai-wei, LIU Yue-ying, LIU Hai-yan, LUO Guang-hong. Cloning and Expression of Protease SpP1 Gene and Characterization of Enzymatic Properties [J]. Biotechnology Bulletin, 2024, 40(4): 278-286. |
| [9] | WANG Lu, LIU Meng-yu, ZHANG Fu-yuan, JI Shou-kun, WANG Yun, ZHANG Ying-jie, DUAN Chun-hui, LIU Yue-qin, YAN Hui. Isolation and Identification of Rumen Skatole-degrading Bacteria and Analysis on Their Degradation Characteristics [J]. Biotechnology Bulletin, 2024, 40(3): 305-311. |
| [10] | CHEN Jin-hang, ZHANG Yi, ZHANG Jun-tao, WEI Ben-mei, WANG Hong-xun, ZHENG Ming-ming. Preparation of Immobilized Lipase for the Solvent-free Synthesis of Cinnamyl Acetate [J]. Biotechnology Bulletin, 2023, 39(9): 97-104. |
| [11] | RAO Zi-huan, XIE Zhi-xiong. Isolation and Identification of a Cellulose-degrading Strain of Olivibacter jilunii and Analysis of Its Degradability [J]. Biotechnology Bulletin, 2023, 39(8): 283-290. |
| [12] | YOU Zi-juan, CHEN Han-lin, DENG Fu-cai. Research Progress in the Extraction and Functional Activities of Bioactive Peptides from Fish Skin [J]. Biotechnology Bulletin, 2023, 39(7): 91-104. |
| [13] | CHE Yong-mei, GUO Yan-ping, LIU Guang-chao, YE Qing, LI Ya-hua, ZHAO Fang-gui, LIU Xin. Isolation and Identification of Bacterial Strain C8 and B4 and Their Halotolerant Growth-promoting Effects and Mechanisms [J]. Biotechnology Bulletin, 2023, 39(5): 276-285. |
| [14] | YANG Jun-zhao, ZHANG Xin-rui, ZHAO Guo-zhu, ZHENG Fei. Structure and Function Analysis of Novel GH5 Multi-domain Cellulase [J]. Biotechnology Bulletin, 2023, 39(4): 71-80. |
| [15] | WANG Feng-ting, WANG Yan, SUN Ying, CUI Wen-jing, QIAO Kai-bin, PAN Hong-yu, LIU Jin-liang. Isolation and Identification of Saline-alkali Tolerant Aspergillus terreus SYAT-1 and Its Activities Against Plant Pathogenic Fungi [J]. Biotechnology Bulletin, 2023, 39(2): 203-210. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||