








Biotechnology Bulletin ›› 2026, Vol. 42 ›› Issue (1): 51-66.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0863
Previous Articles Next Articles
LIU Huan1,2( ), GUO Fa-xu3, ZHAO Xiao-yan2, HUANG Long-yu2, WANG Jian1, ZHOU Guo-min4,5,6(
), GUO Fa-xu3, ZHAO Xiao-yan2, HUANG Long-yu2, WANG Jian1, ZHOU Guo-min4,5,6( ), ZHANG Jian-hua1,2(
), ZHANG Jian-hua1,2( )
)
Received:2025-08-09
Online:2026-01-26
Published:2026-02-04
Contact:
ZHOU Guo-min, ZHANG Jian-hua
E-mail:liuhuan01@139.com;zhouguomin@caas.cn;zhangjianhua@caas.cn
LIU Huan, GUO Fa-xu, ZHAO Xiao-yan, HUANG Long-yu, WANG Jian, ZHOU Guo-min, ZHANG Jian-hua. Advances in Artificial Intelligence for DNA Design[J]. Biotechnology Bulletin, 2026, 42(1): 51-66.
| 模型 Model | 架构类型 Architecture type | 主要用途 Primary applications | 特点 Features | 优势 Advantages | 局限 Limitations |
|---|---|---|---|---|---|
| Enformer | Transformer | 功能预测 | 长程依赖 | 泛化性强 | 数据需求量大 |
| DNABERT | Transformer | 序列特征提取 | k-mer嵌入 | 泛化能力好 | 解释性有限 |
| Evo/Evo2 | Transformer | 序列设计/预测 | 跨模态泛化生 | 零样本迁移 | 复杂度较高 |
| GAN | 对抗生成网络 | 序列设计/预测 | 成创新序列 | 功能多样 | 训练稳定性差 |
| VAE | 自编码器 | 序列优化 | 潜在空间优化 | 控制性强 | 泛化较难 |
| Diffusion | 扩散模型 | 序列精准生成 | 稳定性强 | 可控性强 | 计算资源需求高 |
Table 1 Introduction of conventional and cutting-edge models in the field of DNA design
| 模型 Model | 架构类型 Architecture type | 主要用途 Primary applications | 特点 Features | 优势 Advantages | 局限 Limitations |
|---|---|---|---|---|---|
| Enformer | Transformer | 功能预测 | 长程依赖 | 泛化性强 | 数据需求量大 |
| DNABERT | Transformer | 序列特征提取 | k-mer嵌入 | 泛化能力好 | 解释性有限 |
| Evo/Evo2 | Transformer | 序列设计/预测 | 跨模态泛化生 | 零样本迁移 | 复杂度较高 |
| GAN | 对抗生成网络 | 序列设计/预测 | 成创新序列 | 功能多样 | 训练稳定性差 |
| VAE | 自编码器 | 序列优化 | 潜在空间优化 | 控制性强 | 泛化较难 |
| Diffusion | 扩散模型 | 序列精准生成 | 稳定性强 | 可控性强 | 计算资源需求高 |
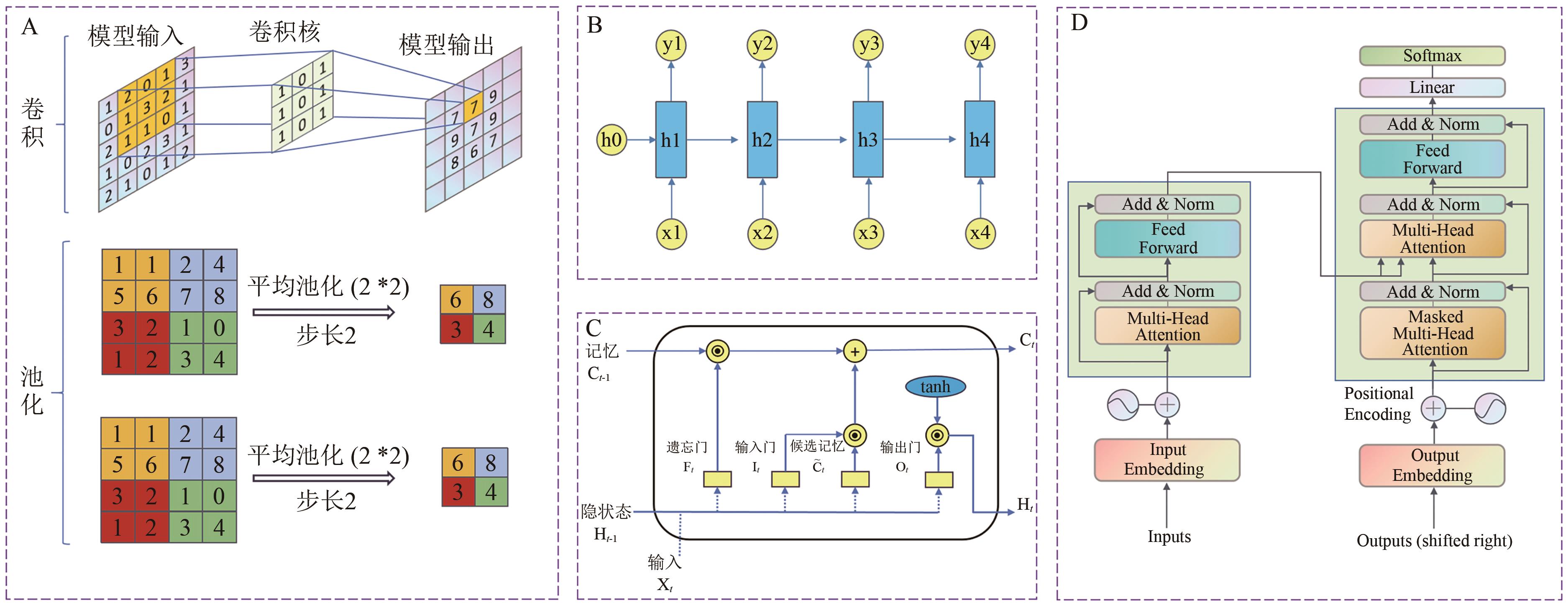
Fig. 2 Architectures of different predictive models, including (A) a Convolutional Neural Network (CNN), (B) a Recurrent Neural Network (RNN), (C) a Long Short-Term Memory (LSTM) network, and (D) a Transformer
数据库/资源 Database/Resource | 内容简介 Description | 类型/范围 Type/Scope | 用途 Applications |
|---|---|---|---|
| GENCODE | 人类注释基因组 | 注释序列 | 基因注释 |
| INSD | 核酸数据集 | 序列库 | 通用序列建模 |
| EPDnew | 启动子信息 | 启动子功能 | 功能预测 |
| EnhancerAtlas | 增强子注释 | 增强子库 | 功能研究 |
| MethBank | 甲基化数据 | 表观遗传 | 调控分析 |
| UCSC Browser | 多组学数据 | 组学集成 | 多组学分析 |
| GEO | 转录组表观组 | 组学数据库 | 转录组分析 |
| ENCODE | 功能元件注释数据 | 多组学 | 功能元件注释 |
| Roadmap | 表观遗传组 | 表观组 | 功能研究 |
| GTEx | 组织表达图谱 | 转录组 | 表达分析 |
| Ensembl | 多物种注释 | 基因组注释 | 跨物种建模 |
| RNAcentral | ncRNA库 | 非编码RNA | 序列分析 |
| GreeNC | 植物ncRNA | 植物数据库 | 功能元件设计 |
| DBHR | 多组学整合 | 综合组学 | 功能研究 |
| UniProt | 蛋白功能库 | 蛋白注释 | 功能预测 |
| MPRA | 高通量实验 | 功能平台 | 功能筛选 |
Table 2 Data used in the field of DNA design, including their sources and applications
数据库/资源 Database/Resource | 内容简介 Description | 类型/范围 Type/Scope | 用途 Applications |
|---|---|---|---|
| GENCODE | 人类注释基因组 | 注释序列 | 基因注释 |
| INSD | 核酸数据集 | 序列库 | 通用序列建模 |
| EPDnew | 启动子信息 | 启动子功能 | 功能预测 |
| EnhancerAtlas | 增强子注释 | 增强子库 | 功能研究 |
| MethBank | 甲基化数据 | 表观遗传 | 调控分析 |
| UCSC Browser | 多组学数据 | 组学集成 | 多组学分析 |
| GEO | 转录组表观组 | 组学数据库 | 转录组分析 |
| ENCODE | 功能元件注释数据 | 多组学 | 功能元件注释 |
| Roadmap | 表观遗传组 | 表观组 | 功能研究 |
| GTEx | 组织表达图谱 | 转录组 | 表达分析 |
| Ensembl | 多物种注释 | 基因组注释 | 跨物种建模 |
| RNAcentral | ncRNA库 | 非编码RNA | 序列分析 |
| GreeNC | 植物ncRNA | 植物数据库 | 功能元件设计 |
| DBHR | 多组学整合 | 综合组学 | 功能研究 |
| UniProt | 蛋白功能库 | 蛋白注释 | 功能预测 |
| MPRA | 高通量实验 | 功能平台 | 功能筛选 |
位置 Position | A密度 A density | C密度 C density | G密度 G density | T密度 T density |
|---|---|---|---|---|
| 1 | 1.00 | 0.00 | 0.00 | 0.00 |
| 2 | 0.50 | 0.00 | 0.50 | 0.00 |
| 3 | 0.33 | 0.00 | 0.33 | 0.33 |
| 4 | 0.25 | 0.25 | 0.25 | 0.25 |
| 5 | 0.20 | 0.20 | 0.20 | 0.10 |
| 6 | 0.17 | 0.17 | 0.33 | 0.33 |
| 7 | 0.14 | 0.29 | 0.29 | 0.29 |
| 8 | 0.25 | 0.25 | 0.25 | 0.25 |
Table 3 Example of ND encoding and the resulting numerical matrix
位置 Position | A密度 A density | C密度 C density | G密度 G density | T密度 T density |
|---|---|---|---|---|
| 1 | 1.00 | 0.00 | 0.00 | 0.00 |
| 2 | 0.50 | 0.00 | 0.50 | 0.00 |
| 3 | 0.33 | 0.00 | 0.33 | 0.33 |
| 4 | 0.25 | 0.25 | 0.25 | 0.25 |
| 5 | 0.20 | 0.20 | 0.20 | 0.10 |
| 6 | 0.17 | 0.17 | 0.33 | 0.33 |
| 7 | 0.14 | 0.29 | 0.29 | 0.29 |
| 8 | 0.25 | 0.25 | 0.25 | 0.25 |
| [1] | Eraslan G, Avsec Ž, Gagneur J, et al. Deep learning: new computational modelling techniques for genomics [J]. Nat Rev Genet, 2019, 20(7): 389-403. |
| [2] | Kelley DR, Reshef YA, Bileschi M, et al. Sequential regulatory activity prediction across chromosomes with convolutional neural networks [J]. Genome Res, 2018, 28(5): 739-750. |
| [3] | Agarwal V, Shendure J. Predicting mRNA abundance directly from genomic sequence using deep convolutional neural networks [J]. Cell Rep, 2020, 31(7): 107663. |
| [4] | Zhang PC, Wang HC, Xu HW, et al. Deep flanking sequence engineering for efficient promoter design using DeepSEED [J]. Nat Commun, 2023, 14: 6309. |
| [5] | Wu MR, Nissim L, Stupp D, et al. A high-throughput screening and computation platform for identifying synthetic promoters with enhanced cell-state specificity (SPECS) [J]. Nat Commun, 2019, 10: 2880. |
| [6] | Yu TC, Liu WL, Brinck MS, et al. Multiplexed characterization of rationally designed promoter architectures deconstructs combinatorial logic for IPTG-inducible systems [J]. Nat Commun, 2021, 12: 325. |
| [7] | Ji YR, Zhou ZH, Liu H, et al. DNABERT: pre-trained Bidirectional Encoder Representations from Transformers model for DNA-language in genome [J]. Bioinformatics, 2021, 37(15): 2112-2120. |
| [8] | Linder J, Bogard N, Rosenberg AB, et al. A generative neural network for maximizing fitness and diversity of synthetic DNA and protein sequences [J]. Cell Syst, 2020, 11(1): 49-62.e16. |
| [9] | Zhou ZH, Ji YR, Li WJ, et al. DNABERT-2: Efficient foundation model and benchmark for multi-species genomes [J]. arXiv.org. 2023. DOI: 10.48550/arxiv.2306.15006 . |
| [10] | Avsec Ž, Weilert M, Shrikumar A, et al. Base-resolution models of transcription-factor binding reveal soft motif syntax [J]. Nat Genet, 2021, 53(3): 354-366. |
| [11] | Wang Y, Wang HC, Wei L, et al. Synthetic promoter design in Escherichia coli based on a deep generative network [J]. Nucleic Acids Res, 2020, 48(12): 6403-6412. |
| [12] | Killoran N, Lee LJ, Delong A, et al. Generating and designing DNA with deep generative models [J]. arXiv.org. 2017. DOI: 10.48550/arxiv.1712.06148 . |
| [13] | Gupta A, Zou J. Feedback GAN for DNA optimizes protein functions [J]. Nat Mach Intell, 2019, 1(2): 105-111. |
| [14] | Wang XL, Xu KJ, Tan YM, et al. Deep learning-assisted design of novel promoters in Escherichia coli [J]. Adv Genet, 2023, 4(4): 2300184. |
| [15] | Alipanahi B, Delong A, Weirauch MT, et al. Predicting the sequence specificities of DNA-and RNA-binding proteins by deep learning [J]. Nat Biotechnol, 2015, 33(8): 831-838. |
| [16] | Wang XL, Xu KJ, Huang ZS, et al. Accelerating promoter identification and design by deep learning [J]. Trends Biotechnol, 2025. |
| [17] | Barbadilla-Martínez L, Klaassen N, van Steensel B, et al. Predicting gene expression from DNA sequence using deep learning models [J]. Nat Rev Genet, 2025, 26(10): 666-680. |
| [18] | Quang D, Xie XH. DanQ: a hybrid convolutional and recurrent deep neural network for quantifying the function of DNA sequences [J]. Nucleic Acids Res, 2016, 44(11): e107. |
| [19] | Jores T, Tonnies J, Wrightsman T, et al. Synthetic promoter designs enabled by a comprehensive analysis of plant core promoters [J]. Nat Plants, 2021, 7(6): 842-855. |
| [20] | Zhou J, Troyanskaya OG. Predicting effects of noncoding variants with deep learning-based sequence model [J]. Nat Meth, 2015, 12(10): 931-934. |
| [21] | Kelley DR, Snoek J, Rinn JL. Basset: learning the regulatory code of the accessible genome with deep convolutional neural networks [J]. Genome Res, 2016, 26(7): 990-999. |
| [22] | Li ZH, Zhang YY, Peng B, et al. A novel interpretable deep learning-based computational framework designed synthetic enhancers with broad cross-species activity [J]. Nucleic Acids Res, 2024, 52(21): 13447-13468. |
| [23] | Yin C, Castillo-Hair S, Byeon GW, et al. Iterative deep learning design of human enhancers exploits condensed sequence grammar to achieve cell-type specificity [J]. Cell Syst, 2025, 16(7): 101302. |
| [24] | Vaishnav ED, de Boer CG, Molinet J, et al. The evolution, evolvability and engineering of gene regulatory DNA [J]. Nature, 2022, 603(7901): 455-463. |
| [25] | Fu HG, Liang YB, Zhong XQ, et al. Codon optimization with deep learning to enhance protein expression [J]. Sci Rep, 2020, 10: 17617. |
| [26] | Yang G, Chen YJ, Guo QH, et al. Leveraging pre-trained AI models for robust promoter sequence design in synthetic biology [J]. Swwlxb, 2025, 11: 1. |
| [27] | Fallahpour A, Gureghian V, Filion GJ, et al. CodonTransformer: a multispecies codon optimizer using context-aware neural networks [J]. Nat Commun, 2025, 16: 3205. |
| [28] | Zrimec J, Fu XZ, Muhammad AS, et al. Controlling gene expression with deep generative design of regulatory DNA [J]. Nat Commun, 2022, 13: 5099. |
| [29] | de Almeida BP, Schaub C, Pagani M, et al. Targeted design of synthetic enhancers for selected tissues in the Drosophila embryo. [J]. Nature, 2024, 626(7997): 207-211. |
| [30] | Avdeyev P, Shi CL, Tan YH, et al. Dirichlet diffusion score model for biological sequence generation [J]. arXiv.org. 2023. DOI: 10.48550/arxiv.2305.10699 . |
| [31] | de Almeida BP, Reiter F, Pagani M, et al. DeepSTARR predicts enhancer activity from DNA sequence and enables the de novo design of synthetic enhancers [J]. Nat Genet, 2022, 54(5): 613-624. |
| [32] | Avsec Ž, Agarwal V, Visentin D, et al. Effective gene expression prediction from sequence by integrating long-range interactions [J]. Nat Meth, 2021, 18(10): 1196-1203. |
| [33] | Yang Y, Lee JH, Poindexter MR, et al. Rational design and testing of abiotic stress-inducible synthetic promoters from poplar cis-regulatory elements [J]. Plant Biotechnol J, 2021, 19(7): 1354-1369. |
| [34] | Jain R, Jain A, Mauro E, et al. ICOR: improving codon optimization with recurrent neural networks [J]. BMC Bioinform, 2023, 24(1): 132. |
| [35] | Lei X, Wang X, Chen GL, et al. Combining diffusion and transformer models for enhanced promoter synthesis and strength prediction in deep learning [J]. mSystems, 2025, 10(4) |
| [36] | Kelley DR. Cross-species regulatory sequence activity prediction [J]. PLoS Comput Biol, 2020, 16(7): e1008050. |
| [37] | Li JQ, Zhang PC, Xi X, et al. Modeling and designing enhancers by introducing and harnessing transcription factor binding units [J]. Nat Commun, 2025, 16: 1469. |
| [38] | Gasperini M, Tome JM, Shendure J. Towards a comprehensive catalogue of validated and target-linked human enhancers [J]. Nat Rev Genet, 2020, 21(5): 292-310. |
| [39] | Zhou J, Theesfeld CL, Yao K, et al. Deep learning sequence-based ab initio prediction of variant effects on expression and disease risk [J]. Nat Genet, 2018, 50(8): 1171-1179. |
| [40] | Friedman RZ, Ramu A, Lichtarge S, et al. Active learning of enhancers and silencers in the developing neural retina [J]. Cell Syst, 2025, 16(1): 101163. |
| [41] | Karbalayghareh A, Sahin M, Leslie CS. Chromatin interaction-aware gene regulatory modeling with graph attention networks [J]. Genome Res, 2022, 32(7): 1290-1304. |
| [42] | Jaganathan K, Kyriazopoulou Panagiotopoulou S, McRae JF, et al. Predicting splicing from primary sequence with deep learning [J]. Cell, 2019, 176(3): 535-548.e24. |
| [43] | Sample PJ, Wang B, Reid DW, et al. Human 5' UTR design and variant effect prediction from a massively parallel translation assay [J]. Nat Biotechnol, 2019, 37(7): 803-809. |
| [44] | Bogard N, Linder J, Rosenberg AB, et al. A deep neural network for predicting and engineering alternative polyadenylation [J]. Cell, 2019, 178(1): 91-106.e23. |
| [45] | Lee NK, Tang ZQ, Toneyan S, et al. EvoAug: improving generalization and interpretability of genomic deep neural networks with evolution-inspired data augmentations [J]. Genome Biol, 2023, 24(1): 105. |
| [46] | Cherednichenko O, Poptsova M. Data augmentation with generative models improves detection of Non-B DNA structures [J]. Comput Biol Med, 2025, 184: 109440. |
| [47] | Davidi D, et al. Regulatory DNA sequence design with reinforcement learning [DB/OL]. arXiv preprint: 2503.07981. |
| [48] | Jaganathan K, Ersaro N, Novakovsky G, et al. Predicting expression-altering promoter mutations with deep learning [J]. Science, 2025, 389(6760): eads7373. |
| [49] | Dalla-Torre H, Gonzalez L, Mendoza-Revilla J, et al. Nucleotide Transformer: building and evaluating robust foundation models for human genomics [J]. Nat Meth, 2025, 22(2): 287-297. |
| [50] | 张冀东, 王志晗, 刘博, 等. 深度学习在生物序列分析领域的应用进展 [J]. 北京工业大学学报, 2022, 48(8): 878-887. |
| Zhang JD, Wang ZH, Liu B, et al. Progress in the applications of deep learning in biological sequence analysis [J]. J Beijing Univ Technol, 2022, 48(8): 878-887. |
| [1] | CAI Ru-feng, YANG Yu-xuan, YU Ji-zheng, LI Jia-nan. Artificial Intelligence Transforms Protein Engineering: From Structural Analysis to Synthetic Biology through Algorithmic Advancements [J]. Biotechnology Bulletin, 2025, 41(8): 1-10. |
| [2] | WANG Hui, FAN Ling-xi, SUN Ji-lu, WANG Yuan, WU Ning-feng, TIAN Jian, GUAN Fei-fei. Enhancing the Thermostability of Lysozyme RPL187 Based on Protein Intelligence Models [J]. Biotechnology Bulletin, 2025, 41(7): 336-346. |
| [3] | GUO Fa-xu, FENG Quan, ZHANG Jian-hua, ZHOU Huan-bin, YANG Sen, WANG Jian, ZHOU Guo-min. Research Advances in AI-driven Enzyme Modifying and Design [J]. Biotechnology Bulletin, 2025, 41(12): 50-65. |
| [4] | HE Yuan, MOU Qiang, HE Yu-bing, ZHAO Xiao-yan, WANG Jian, ZHOU Guo-min, ZHANG Jian-hua. Advances in Protein Mining and Design Based on Artificial Intelligence [J]. Biotechnology Bulletin, 2025, 41(10): 143-155. |
| [5] | JI Hong-chao, LI Zheng-yan. Research Progress and Prospects in the Structural Annotation of Unknown Secondary Metabolites Based on Mass Spectrometry [J]. Biotechnology Bulletin, 2024, 40(10): 76-85. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||