








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (11): 285-292.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0101
Previous Articles Next Articles
FENG Min1( ), LI Shu-ting2, ZHANG Yang-zi2, SU Yuan2, ZHU Long-jiao2, CAO Ji-juan3, LIU Hai-yan1(
), LI Shu-ting2, ZHANG Yang-zi2, SU Yuan2, ZHU Long-jiao2, CAO Ji-juan3, LIU Hai-yan1( ), XU Wen-tao2(
), XU Wen-tao2( )
)
Received:2021-01-27
Online:2021-11-26
Published:2021-12-03
Contact:
LIU Hai-yan,XU Wen-tao
E-mail:981838423@qq.com;freeair772000@163.com;xuwentao@cau.edu.cn
FENG Min, LI Shu-ting, ZHANG Yang-zi, SU Yuan, ZHU Long-jiao, CAO Ji-juan, LIU Hai-yan, XU Wen-tao. Development of a Innovative Fluorescent Quantitative PCR Method for Salmonella Based on Fluorescent Self-quenching Primers[J]. Biotechnology Bulletin, 2021, 37(11): 285-292.
| 名称Name | 序列Sequence(5'-3') | 碱基数Number of bases/nt |
|---|---|---|
| 上游引物Forward primer(FP) | CGGGTCAAGGCTGAGGAA | 18 |
| 原始下游引物Original reverse primer(ORP) | TGCTGAAGTTGAGGATGTTATTCG | 24 |
| 下游引物Reverse primer(RP) 靶基因Target gene | CGAATATGCTGAAGTTGAGGATGTTATT(FAM)CG TGCTGAAGTTGAGGATGTTATTCGCAAAGGGATCCGTCA GACCTCTGGCAGTACCTTCCTCAGCCTTGACCCG | 30 85 |
Table 1 Primer and target gene sequence
| 名称Name | 序列Sequence(5'-3') | 碱基数Number of bases/nt |
|---|---|---|
| 上游引物Forward primer(FP) | CGGGTCAAGGCTGAGGAA | 18 |
| 原始下游引物Original reverse primer(ORP) | TGCTGAAGTTGAGGATGTTATTCG | 24 |
| 下游引物Reverse primer(RP) 靶基因Target gene | CGAATATGCTGAAGTTGAGGATGTTATT(FAM)CG TGCTGAAGTTGAGGATGTTATTCGCAAAGGGATCCGTCA GACCTCTGGCAGTACCTTCCTCAGCCTTGACCCG | 30 85 |
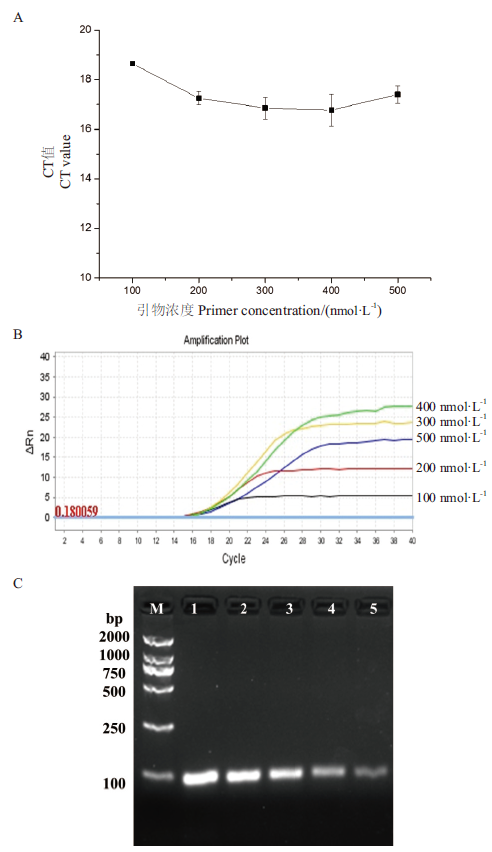
Fig.2 Primer concentration optimization A: CT value comparison chart. B: Amplification curve. C: Agarose gel electrophoresis chart. Lanes 1-5 are amplified product at primer concentrations of 500 nmol/L, 400 nmol/L, 300 nmol/L, 200 nmol/L, 100 nmol/L, respectively
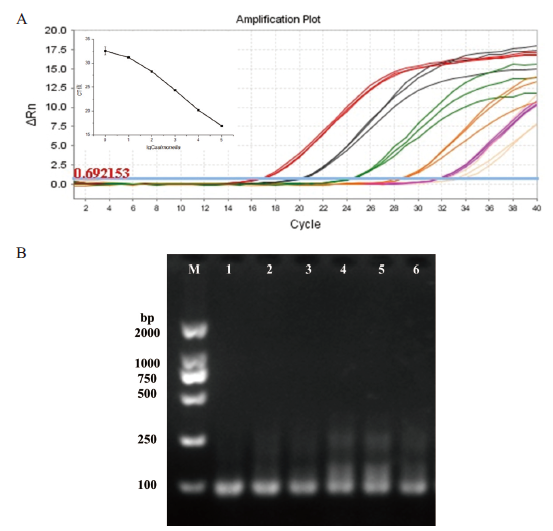
Fig.3 Sensitivity detection A: Amplification curve: the Salmonella concentrations corresponding to the big red line, black line, green line, orange line, purple line and pink line are 105 CFU/mL, 104 CFU/mL、103 CFU/mL, 102 CFU/mL, 101 CFU/mL and 100 CFU/mL (the attached shows the relationship between the CT value corresponding to the amplification curve and lgCsalmonella); B: agarose gel electrophoresis diagram: the concentration of Salmonella corresponding to lanes 1-6 is 105 CFU/mL, 104 CFU/mL, 103 CFU/mL, 102 CFU/mL, 101 CFU/mL and 100 CFU/mL
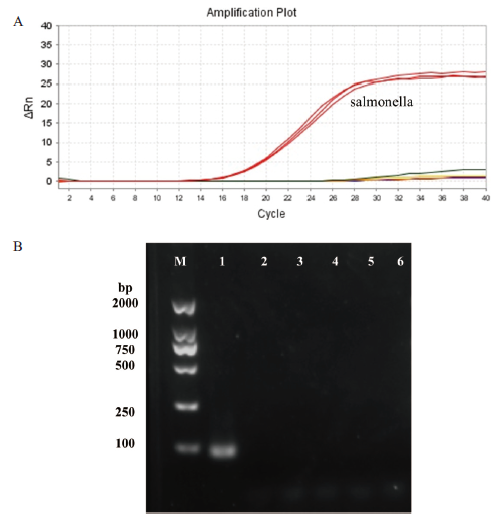
Fig.5 Specific detection A: Amplification curve: The food-borne pathogen corresponding to the big red line is Salmonella, and the other color lines represent Staphylococcus aureus, Bacillus cereus, Escherichia coli, Listeria monocytogenes, and Vibrio parahaemolyticus. B :Agarose gel electrophoresis diagram: lanes 1-6 are the products of DNA amplification of Salmonella, Staphylococcus aureus, Bacillus cereus, Escherichia coli, Listeria monocytogenes, and Vibrio parahaemolyticus, respectively
| 沙门氏菌的浓度 Concentration of Salmonella/(CFU·mL-1) | CT值 CT value | ±S | RSD | ||
|---|---|---|---|---|---|
| 105 | 16.9 | 16.98 | 16.89 | 16.9±0.05 | 0.29% |
| 104 | 20.26 | 20.29 | 20.18 | 20.2±0.05 | 0.28% |
| 103 | 23.66 | 23.63 | 23.55 | 23.6±0.06 | 0.24% |
| 102 | 26.49 | 26.43 | 26.34 | 26.4±0.08 | 0.29% |
Table 2 IFQ-PCR method for the detection results of different concentrations of Salmonella in milk samples
| 沙门氏菌的浓度 Concentration of Salmonella/(CFU·mL-1) | CT值 CT value | ±S | RSD | ||
|---|---|---|---|---|---|
| 105 | 16.9 | 16.98 | 16.89 | 16.9±0.05 | 0.29% |
| 104 | 20.26 | 20.29 | 20.18 | 20.2±0.05 | 0.28% |
| 103 | 23.66 | 23.63 | 23.55 | 23.6±0.06 | 0.24% |
| 102 | 26.49 | 26.43 | 26.34 | 26.4±0.08 | 0.29% |
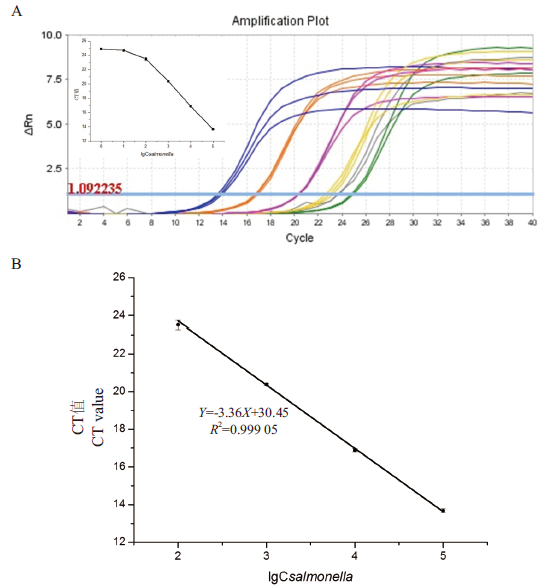
Fig.7 Sensitivity detection of FQ-PCR based on dye method A: Amplification curve: The Salmonella concentrations corresponding to the blue line, orange line, purple line, yellow line, gray line, and green line are 105 CFU/mL, 104 CFU/mL, 103 CFU/mL, 102 CFU/mL, 101 CFU/mL and 100 CFU/mL (The attached shows the relationship between the CT value corresponding to the amplification curve and lgCsalmonella). B: FQ-PCR method standard curve
| 方法Method | 优点Advantage | 缺点Disadvantage |
|---|---|---|
| IFQ-PCR法 IFQ-PCR method | 引物的修饰简单、合成成本相对较低、扩增体系简单、可减少引物二聚体和碱基错配的发生 | 引物的设计要求高 |
| FQ-PCR染料法 FQ-PCR dye method | 无需标记、灵敏度高、成本低、可通过熔解曲线分析结果 | 特异性差、扩增体系需额外添加染料 |
| 探针法 Probe method | 特异性最强、不受非特异性扩增与引物二聚体的影响 | 探针的设计复杂、标记成本高、扩增体系复杂 |
Table 3 Comparison of IFQ-PCR method and FQ-PCR method [13-16, 19-23]
| 方法Method | 优点Advantage | 缺点Disadvantage |
|---|---|---|
| IFQ-PCR法 IFQ-PCR method | 引物的修饰简单、合成成本相对较低、扩增体系简单、可减少引物二聚体和碱基错配的发生 | 引物的设计要求高 |
| FQ-PCR染料法 FQ-PCR dye method | 无需标记、灵敏度高、成本低、可通过熔解曲线分析结果 | 特异性差、扩增体系需额外添加染料 |
| 探针法 Probe method | 特异性最强、不受非特异性扩增与引物二聚体的影响 | 探针的设计复杂、标记成本高、扩增体系复杂 |
| [1] |
Hao LL, Gu HJ, et al. A chemiluminescent aptasensor based on rolling circle amplification and Co2+/N-(aminobutyl)-N-(ethylisoluminol)functional flowerlike gold nanoparticles for Salmonella typhimurium detection[J]. Talanta, 2017, 164: 275-282.
doi: 10.1016/j.talanta.2016.11.053 URL |
| [2] |
Scharff RL. Economic burden from health losses due to foodborne illness in the United States[J]. J Food Prot, 2012, 75(1): 123-131.
doi: 10.4315/0362-028X.JFP-11-058 URL |
| [3] |
Wang L, et al. QCM-based aptamer selection and detection of Salmonella typhimurium[J]. Food Chem, 2017, 221: 776-782.
doi: 10.1016/j.foodchem.2016.11.104 URL |
| [4] |
Li ST, et al. Luminescent DNAzyme and universal blocking linker super polymerase chain reaction visual biosensor for the detection of Salmonella[J]. Food Chem, 2020, 324: 126859.
doi: 10.1016/j.foodchem.2020.126859 URL |
| [5] |
Paniel N, Baudart J, Hayat A, et al. Aptasensor and genosensor methods for detection of microbes in real world samples[J]. Methods, 2013, 64(3): 229-240.
doi: 10.1016/j.ymeth.2013.07.001 pmid: 23872322 |
| [6] |
Narmani A, Rezvani M, Farhood B, et al. Folic acid functionalized nanoparticles as pharmaceutical carriers in drug delivery systems[J]. Drug Dev Res, 2019, 80(4): 404-424.
doi: 10.1002/ddr.v80.4 URL |
| [7] | 孙晓飞, 赵凯, 王金斌, 等. 食源性病原菌检测方法研究进展[J]. 现代农业科技, 2010(14): 334-336. |
| Sun XF, Zhao K, Wang JB, et al. Research progress of detection technology in foodborne pathogens[J]. Mod Agric Sci Technol, 2010(14): 334-336. | |
| [8] | 张冲, 刘祥, 陈计峦. 实时荧光定量RT-PCR检测沙门氏菌活菌[J]. 食品工业科技, 2012, 33(6): 91-94. |
| Zhang C, Liu X, Chen JL. Real-time quantitative reverse transcription polymerase chain reaction detection of live Salmonella[J]. Sci Technol Food Ind, 2012, 33(6): 91-94. | |
| [9] | 王凤军, 叶素丹. TaqMan实时荧光PCR对沙门氏菌能力验证样品的快速检测与鉴定[J]. 现代食品科技, 2020, 36(12): 300-306, 83. |
| Wang FJ, Ye SD. Rapid detection and identification of Salmonella in proficiency testing samples by TaqMan real-time fluorescent PCR[J]. Mod Food Sci Technol, 2020, 36(12): 300-306, 83. | |
| [10] | Singh LP, Yadav AS, Singh RP, et al. Detection and quantification of Salmonella on chicken egg shell by real time PCR[J]. Indian J Animal Sci, 2013, 83(5): 481-483. |
| [11] |
Lee N, Kwon KY, Oh SK, et al. A multiplex pcr assay for simultaneous detection of Escherichia coli O157:H7, Bacillus cereus, Vibrio parahaemolyticus, Salmonella spp. Listeria monocytogenes, and Staphylococcus aureus in Korean Ready-to-Eat food[J]. Foodborne Pathog Dis, 2014, 11(7): 574-580.
doi: 10.1089/fpd.2013.1638 URL |
| [12] |
Rodríguez A, Rodríguez M, Córdoba JJ, et al. Design of primers and probes for quantitative real-time PCR methods[J]. Methods Mol Biol, 2015, 1275: 31-56.
doi: 10.1007/978-1-4939-2365-6_3 pmid: 25697650 |
| [13] |
Nazarenko I, Lowe B, Darfler M, et al. Multiplex quantitative PCR using self-quenched primers labeled with a single fluorophore[J]. Nucleic Acids Res, 2002, 30(9): e37.
doi: 10.1093/nar/30.9.e37 URL |
| [14] |
Crockett AO, Wittwer CT. Fluorescein-labeled oligonucleotides for real-time pcr:using the inherent quenching of deoxyguanosine nucleotides[J]. Anal Biochem, 2001, 290(1): 89-97.
pmid: 11180941 |
| [15] |
Nazarenko I, et al. Effect of primary and secondary structure of oligodeoxyribonucleotides on the fluorescent properties of conjugated dyes[J]. Nucleic Acids Res, 2002, 30(9): 2089-2195.
pmid: 11972350 |
| [16] |
Knemeyer JP, Marmé N, Sauer M. Probes for detection of specific DNA sequences at the single-molecule level[J]. Anal Chem, 2000, 72(16): 3717-3724.
pmid: 10959954 |
| [17] |
Zhou ZQ, Zhang YZ, Guo MZ, et al. Ultrasensitive magnetic DNAzyme-copper nanoclusters fluorescent biosensor with triple amplification for the visual detection of E. coli O157:H7[J]. Biosens Bioelectron, 2020, 167: 112475.
doi: 10.1016/j.bios.2020.112475 URL |
| [18] |
Hui CY, Liu M, Li YF, et al. A paper sensor printed with multifunctional bio/nano materials[J]. Angew Chem Int Ed, 2018, 57(17): 4549-4553.
doi: 10.1002/anie.v57.17 URL |
| [19] |
Rani N, Vajpayee P, Bhatti S, et al. Quantification of Salmonella Typhi in water and sediments by molecular-beacon based qPCR[J]. Ecotoxicol Environ Saf, 2014, 108: 58-64.
doi: 10.1016/j.ecoenv.2014.06.033 URL |
| [20] |
Li B, Chen JQ. Development of a sensitive and specific qPCR assay in conjunction with propidium monoazide for enhanced detection of live Salmonella spp. in food[J]. BMC Microbiol, 2013, 13: 273.
doi: 10.1186/1471-2180-13-273 URL |
| [21] |
Kim SA, Park SH, Lee SI, et al. Development of a rapid method to quantify Salmonella Typhimurium using a combination of MPN with qPCR and a shortened time incubation[J]. Food Microbiol, 2017, 65: 7-18.
doi: 10.1016/j.fm.2017.01.013 URL |
| [22] |
Richards AK, Hopkins BA, Shariat NW. Conserved CRISPR arrays in Salmonella enterica serovar Infantis can serve as qPCR targets to detect Infantis in mixed serovar populations[J]. Lett Appl Microbiol, 2020, 71(2): 138-145.
doi: 10.1111/lam.v71.2 URL |
| [23] |
Azinheiro S, Carvalho J, Prado M, et al. Multiplex detection of Salmonella spp. E. coli O157 and L. monocytogenes by qPCR melt curve analysis in spiked infant formula[J]. Microorganisms, 2020, 8(9): 1359.
doi: 10.3390/microorganisms8091359 URL |
| [1] | YU Hui, WANG Jing, LIANG Xin-xin, XIN Ya-ping, ZHOU Jun, ZHAO Hui-jun. Isolation and Functional Verification of Genes Responding to Iron and Cadmium Stresses in Lycium barbarum [J]. Biotechnology Bulletin, 2023, 39(7): 195-205. |
| [2] | LI Hui-jie, DONG Lian-hua, CHEN Gui-fang, LIU Si-yuan, YANG Jia-yi, YANG Jing-ya. Establishment of Droplet Digital PCR Assay for Quantitative Detection of Pseudomonas cocovenenans in Foods [J]. Biotechnology Bulletin, 2023, 39(1): 127-136. |
| [3] | LIU Li-hui, CHU Jin-hua, SUI Yu-xin, CHEN Yang, CHENG Gu-yue. Research Progress of Main Virulence Factors in Salmonella [J]. Biotechnology Bulletin, 2022, 38(9): 72-83. |
| [4] | LAN Xin-yue, LIU Ning-ning, ZHU Long-jiao, CHEN Xu, CHU Hua-shuo, LI Xiang-yang, DUAN Nuo, XU Wen-tao. Tetracycline Bivalent Aptamer Non-enzyme Label-free Sensor [J]. Biotechnology Bulletin, 2022, 38(3): 276-284. |
| [5] | XIAO Bing, LUO Yun-bo, HUANG Kun-lun, ZHANG Yuan, XU Wen-tao. Research Progress in the Quantitative and Unitive Detecting Technologies Based on Functional Nucleic Acid and Labeled Fluorescence [J]. Biotechnology Bulletin, 2019, 35(7): 213-221. |
| [6] | XIAO Bing, LIU Bang, LUO Yun-bo, HUANG Kun-lun, ZHANG Yuan, LI Xia-ying, ZHANG Xiu-jie, XU Wen-tao, ZHOU Xiang. Research Progress in Quantitative and Unitive Detecting Technologies of Functional Nucleic Acid and Label-Free Fluorescence [J]. Biotechnology Bulletin, 2019, 35(3): 194-202. |
| [7] | GAO Zhi-qiang, WANG Lin, PU Jing, YIN Yi, ZHANG Wei, ZHAO Xiang-peng, YAO Zhen-yu. Duplex Real-time PCR Methods for Quantitative Detection of Bovine Derived Materials in Animal Products [J]. Biotechnology Bulletin, 2018, 34(9): 190-194. |
| [8] | HE Ting-ting, SONG Ting, WANG Chao, ZHANG Chang-bin, WANG Hai-yan. Screening of Reference Genes in Bacillus pumilus by Real-time Fluorescence Quantitative PCR [J]. Biotechnology Bulletin, 2016, 32(11): 99-106. |
| [9] | Wang Fengjun, Liu Shasha, Feng Junli. Rapid Identification of Four Kinds of Decay Fruit Moths by Real-time Fluorescence Quantitative PCR [J]. Biotechnology Bulletin, 2015, 31(2): 78-83. |
| [10] | Wei Di, Gao Jing, Chi Zhenfen, Zhang Guirong, Nie Lingyun. A Novel Method for Detecting Activity of Mitochondria-targeted Nuclease [J]. Biotechnology Bulletin, 2015, 31(12): 81-90. |
| [11] | Li Weiwei, Dun Baoqing, Wang Zhi, Qu Juanjuan. Modified Method for the Efficient and Fast Extraction of Total RNAfrom Saccharomyces cerevisiae with Hot-Phenol [J]. Biotechnology Bulletin, 2012, 0(12): 163-166. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||