








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (5): 248-258.doi: 10.13560/j.cnki.biotech.bull.1985.2020-1206
Previous Articles Next Articles
FENG Cui-lian1( ), WAN Yue2, WANG Jun-gang1, FENG Xiao-yan1, ZHAO Ting-ting1, WANG Wen-zhi1, SHEN Lin-bo1, ZHANG Shu-zhen1,2(
), WAN Yue2, WANG Jun-gang1, FENG Xiao-yan1, ZHAO Ting-ting1, WANG Wen-zhi1, SHEN Lin-bo1, ZHANG Shu-zhen1,2( )
)
Received:2020-09-24
Online:2021-05-26
Published:2021-06-11
Contact:
ZHANG Shu-zhen
E-mail:fengcuilian @itbb.org.cn;zhangshuzhen@itbb.org.cn
FENG Cui-lian, WAN Yue, WANG Jun-gang, FENG Xiao-yan, ZHAO Ting-ting, WANG Wen-zhi, SHEN Lin-bo, ZHANG Shu-zhen. Establishment of a Transformant-specific Detection Method for Cry1Ac-2A-gna Transgenic Sugarcane BCG-17[J]. Biotechnology Bulletin, 2021, 37(5): 248-258.
| 探针 Probe | 引物 Primer | 引物序列 Primer sequence(5'-3') | 退火温度Tm/℃ | 探针长度 Probe length/bp |
|---|---|---|---|---|
| BG | BG-F | CCTCGTTAGACTCAACAGCA | 58 | 857 |
| BG-R | GCTTTATCTTTCCAGCAGTAG | |||
| bar | bar2S | CATCGTCAACCACTACATCGAGACAAGC | 60 | 450 |
| bar3A | CCACTACATCGAGACAAGCACGGTCAAC | |||
| sta | sta-F | GCCGCCGTCTAAAAAGGTGATG | 60 | 682 |
| sta-R | GTCGCCCTCGGGTTCTGATTC | |||
| kan | kan-F | ATCGAAAAATACCGCTGCGTAAAAG | 56 | 637 |
| kan-R | GGACGCAGAAGGCAATGTCATAC |
Table 1 Primers used for PCR DIG probe preparation
| 探针 Probe | 引物 Primer | 引物序列 Primer sequence(5'-3') | 退火温度Tm/℃ | 探针长度 Probe length/bp |
|---|---|---|---|---|
| BG | BG-F | CCTCGTTAGACTCAACAGCA | 58 | 857 |
| BG-R | GCTTTATCTTTCCAGCAGTAG | |||
| bar | bar2S | CATCGTCAACCACTACATCGAGACAAGC | 60 | 450 |
| bar3A | CCACTACATCGAGACAAGCACGGTCAAC | |||
| sta | sta-F | GCCGCCGTCTAAAAAGGTGATG | 60 | 682 |
| sta-R | GTCGCCCTCGGGTTCTGATTC | |||
| kan | kan-F | ATCGAAAAATACCGCTGCGTAAAAG | 56 | 637 |
| kan-R | GGACGCAGAAGGCAATGTCATAC |
| 名称 Name | 引物序列(5'-3') Sequence(5'-3') | Tm/℃ |
|---|---|---|
| Lsp1 | CCATCGTCAACCACTACATCGAGACA | 62 |
| Lsp2 | GCCCCTGGAAGGCACGCAA | 66 |
| Lsp3 | CGTGGTCGCTGTCATCGGGCT | 65 |
| Lsp4 | CTTCAAGCACGGGAACTGGCAT | 64 |
| Rsp1 | GCCCTAAAAAGATAAAACTGTAGAGTCCTGT | 64 |
| Rsp2 | GCGCTTTATATGATTCTCTAAAACACTGATAT | 60 |
| Rsp3 | CTTCAAACAAGTGTGACAAAAAAAATATGT | 59 |
| Rsp4 | GTGACTGGGAAAACCCTGGCGTT | 62 |
Table 2 Specific primers required for nested PCR reactions
| 名称 Name | 引物序列(5'-3') Sequence(5'-3') | Tm/℃ |
|---|---|---|
| Lsp1 | CCATCGTCAACCACTACATCGAGACA | 62 |
| Lsp2 | GCCCCTGGAAGGCACGCAA | 66 |
| Lsp3 | CGTGGTCGCTGTCATCGGGCT | 65 |
| Lsp4 | CTTCAAGCACGGGAACTGGCAT | 64 |
| Rsp1 | GCCCTAAAAAGATAAAACTGTAGAGTCCTGT | 64 |
| Rsp2 | GCGCTTTATATGATTCTCTAAAACACTGATAT | 60 |
| Rsp3 | CTTCAAACAAGTGTGACAAAAAAAATATGT | 59 |
| Rsp4 | GTGACTGGGAAAACCCTGGCGTT | 62 |
| 名称 Name | 引物序列 Primer sequence(5'-3’) | Tm/℃ | 产物长度 Fragment length/bp |
|---|---|---|---|
| LS132 LA797 | CGCTCATGTGTTGAGCATATAAGAAA | 58 | 665 |
| GCAATCCCACGGACCCACAA | |||
| LS132 LA791 | CGCTCATGTGTTGAGCATATAAGAAA | 60 | 659 |
| CCACGGACCCACAACTGTCTTTT | |||
| LS040 LA791 | GTCCTGCCCGTCACCGAGATTT | 61 | 751 |
| CCACGGACCCACAACTGTCTTTT | |||
| RS139 RA467 | GTGACTGGGAAAACCCTGGCGTT | 56 | 328 |
| CTGATGGAAGAGTGGTCAAAAGATGT | |||
| RS139 RA526 | GTGACTGGGAAAACCCTGGCGTT | 57 | 387 |
| GCTTCACACTTGTTGGACAGGATTT | |||
| RS006 RA467 | CTTCAAACAAGTGTGACAAAAAAAATATGT | 55 | 461 |
| CTGATGGAAGAGTGGTCAAAAGATGT |
Table 3 Primers for BCG-17 event-Specific PCR detection
| 名称 Name | 引物序列 Primer sequence(5'-3’) | Tm/℃ | 产物长度 Fragment length/bp |
|---|---|---|---|
| LS132 LA797 | CGCTCATGTGTTGAGCATATAAGAAA | 58 | 665 |
| GCAATCCCACGGACCCACAA | |||
| LS132 LA791 | CGCTCATGTGTTGAGCATATAAGAAA | 60 | 659 |
| CCACGGACCCACAACTGTCTTTT | |||
| LS040 LA791 | GTCCTGCCCGTCACCGAGATTT | 61 | 751 |
| CCACGGACCCACAACTGTCTTTT | |||
| RS139 RA467 | GTGACTGGGAAAACCCTGGCGTT | 56 | 328 |
| CTGATGGAAGAGTGGTCAAAAGATGT | |||
| RS139 RA526 | GTGACTGGGAAAACCCTGGCGTT | 57 | 387 |
| GCTTCACACTTGTTGGACAGGATTT | |||
| RS006 RA467 | CTTCAAACAAGTGTGACAAAAAAAATATGT | 55 | 461 |
| CTGATGGAAGAGTGGTCAAAAGATGT |
| 探针 Probe | BCG-17 | pNUBG | ||
|---|---|---|---|---|
| 限制性内切酶 Restriction enzyme | 杂交带长度 Hybrid band length/bp | 限制性内切酶 Restriction enzyme | 杂交带长度 Hybrid band length/bp | |
| BG | BamHI | 2300 | Kpn I | 11225 |
| Nde I | ≥4650 | |||
| bar | BamHI | 1830 | Kpn I | 1823 |
| EcoRV | ≥1000 | |||
| sta | Kpn I | / | Kpn I | 11225 |
| kan | Kpn I | / | Kpn I | 11225 |
Table 4 Theoretical values of Southern Blot hybridization zone size in BCG-17 and pNUBG
| 探针 Probe | BCG-17 | pNUBG | ||
|---|---|---|---|---|
| 限制性内切酶 Restriction enzyme | 杂交带长度 Hybrid band length/bp | 限制性内切酶 Restriction enzyme | 杂交带长度 Hybrid band length/bp | |
| BG | BamHI | 2300 | Kpn I | 11225 |
| Nde I | ≥4650 | |||
| bar | BamHI | 1830 | Kpn I | 1823 |
| EcoRV | ≥1000 | |||
| sta | Kpn I | / | Kpn I | 11225 |
| kan | Kpn I | / | Kpn I | 11225 |
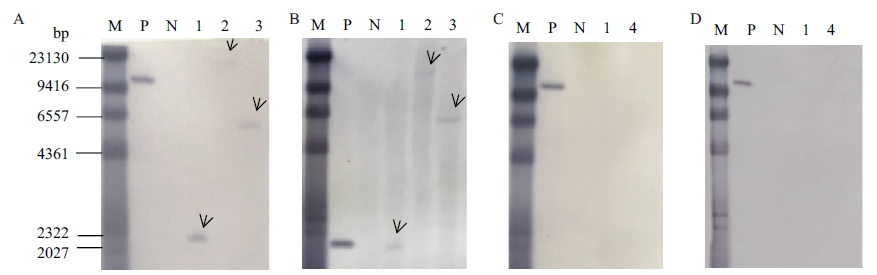
Fig. 3 Southern Blot analysis of transgenic sugarcane BCG-17 A: BG prob. B: Bar probe. C:sta probe. D: kan probe. M: DNA marker for southern blot, P: Kpn I enzyme digest plasmid, N: Nde I enzyme digest Non-GM sugarcane genome, 1: BamHI enzyme digestion, 2: EcoRⅤ enzyme digestion, 3: Nde I enzyme digestion, 4: Hind Ⅲ enzyme digestion
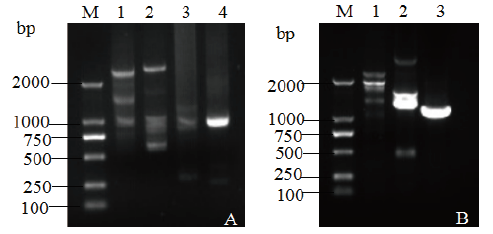
Fig. 4 Nested PCR amplification of left and right flank sequence of BCG-17 A: Left flanking sequence. B: Right flanking sequence. M:DNA marker,1: Amplification for 1st, 2: Amplification for 2nd, 3: Amplification for 3rd,4: Amplification for 4th

Fig. 5 Left flanking sequence of BCG-17 and the character of junction region A: The left flanking sequence of BCG-17. The underlined sequences are T-DNA (511-840 bp),the rest are the sugarcane genome sequence. B: The character of junction region. pUNBG: vector sequences including the left border of T-DNA. The arrow, the break position when the T-DNA was transferred into the sugarcane genome, the left side of the arrow is non-T-DNA region, the red sequences,the core sequence of left T-DNA border, the wavy line, the missing sequence when the T-DNA was transferred into the sugarcane genome, lowercase letters, the left flanking sequence of BCG-17 sugarcane genome, square, overlapping vector sequence of BCG-17 and pUNBG

Fig. 6 Right flanking sequence of BCG-17 and the character of junction region A: The right flanking sequence of BCG-17. The underlined sequences are T-DNA (1-318 bp),the rest are the sugarcane genome sequence. B: The character of junction region. pUNBG: vector sequences including the right border of T-DNA. The arrow: the break position when the T-DNA was transferred into the sugarcane genome, the right side of the arrow is non-T-DNA region, the red sequences, the core sequence of right T-DNA border. The wavy line: the missing sequence when the T-DNA was transferred into the sugarcane genome. BCG-17: The obtained right flanking sequence;lowercase letters, sugarcane genome, square,overlapping vector sequence of BCG-17 and pUNBG
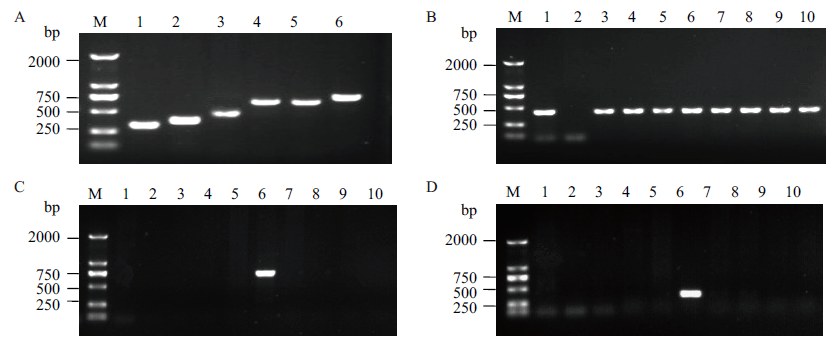
Fig. 7 Transformation event-specific PCR detection of the BCG-17 sugarcane 170A: Screening of event-specific primers for left and right flanking. M: DNA marker. 1-3: PCR products amplified by right primers RS139/RA467, RS139/RA526, RS006/RA467. 4-6: PCR products amplified by left primersLS132/LA797, LS132/LA791, LS040/LA791. B: PCR detection of bar gene in transgenic sugarcane transformed with Cry1c-2A-gna gene. C: Event-specific PCR detection of the left flanking in transgenic sugarcane transformed with Cry1c-2A-gna gene(primer pairs: LS040/LA791 ). D: Event-specific PCR detection of the right flanking in transgenic sugarcane transformed with Cry1c-2A-gna gene(primer pairs:RS139/RA526). M: DNA marker. 1: Expression vector plasmid pUNBG. 2: Non-GM sugarcane ROC22. 3-10:transgenic sugarcane transformed with Cry1c-2A-gna gene BCG-1, BCG-2, BCG-4, BCG-17, BCG-32, BCG-35, BCG-36和BCG-41 respectively
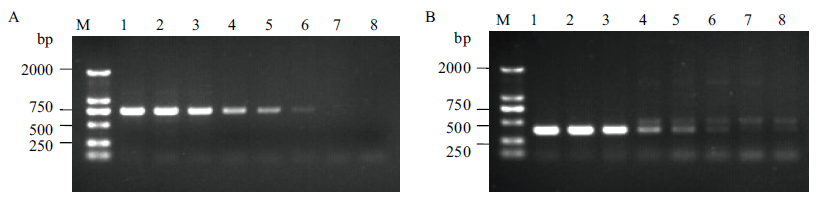
Fig. 8 Limitation of event-specific detection by use of specific primer pairs of left and right flanking sequences A: Limitation of event-specific detection with primer pairs of left flanking(LS040/LA791). B: Limitation of event-specific detection with primer pairs of right flanking(RS139/RA526). M: DNA marker. 1-8: The DNA content of transgenic sugarcane BCG-17 is 100 %, 50 %, 10 %, 1 %, 0.5 %, 0.1 %, 0.05 % and 0 % (non-GM sugarcane ROC22), respectively
| [1] | http://cn.agropages.com/News/NewsDetail---20390.htm. |
| [2] | 祁洋, 李燕, 王永智, 等. 扩增T-DNA插入位点侧翼序列的方法及其应用进展[J]. 安徽农业科学, 2009,37(17):7907-7908, 7927. |
| Qi Y, Li Y, Wang YZ, et al. Advances in the amplification methods and application of T-DNA insertion site flanking sequence[J]. Journal of Anhui Agricultural Sciences, 2009,37(17):7907-7908, 7927. | |
| [3] | 蒲远波, 郭翠, 平淑珍. 基因侧翼序列扩增技术的应用进展[J]. 生物技术通报, 2016,32(11):72-79. |
| Pu YB, Guo C, Ping SZ. Application progress on amplified methods to clone flanking sequence[J]. Biotechnology Bulletin, 2016,32(11):72-79. | |
| [4] | 李付鹏, 伍宝朵, 马朝芝, 等. 基于PCR的染色体步移技术研究进展[J]. 中国生物工程杂志, 2010,30(12):87-94. |
| Li FP, Wu BD, Ma CZ, et al. Progress of chromosome walking by PCR amplification techniques[J]. China Biotechnology, 2010,30(12):87-94. | |
| [5] | 康丹, 方小艳, 游腾飞, 等. 染色体步移技术克隆已知序列侧翼启动子的研究进展[J]. 农业生物技术学报, 2013,21(3):355-366. |
| Kang D, Fang XY, You TF, et al. Progress of chromosome walking the unknown promoter sequences flanked by a known segment[J]. Journal of Agricultural Biotechnology, 2013,21(3):355-366. | |
| [6] | 金永梅, 马瑞, 于志晶, 等. 转基因水稻吉生粳2号的外源基因旁侧序列分离及事件特异性PCR检测方法[J]. 东北农业科学, 2016,41(1):14-19. |
| Jin YM, Ma Rui, Yu ZJ, et al. Identification of the T-DNA flanking sequences and event-specific PCR detection of transgenic rice‘Jishengjing 2’[J]. Journal of Northeast Agricultural Sciences, 2016,41(1):14-19. | |
| [7] |
Dong Y, Jin X, Tang Q, et al. Development and event-specific detection of transgenic glyphosate-resistant rice expressing the G2-EPSPS gene[J], Front Plant Sci, 2017,8:885-895.
doi: 10.3389/fpls.2017.00885 URL |
| [8] | 崔帅, 王作平, 于江辉, 等. 转基因水稻BPL9K-2事件特异性检测方法的建立[J]. 中国生物工程杂志, 2018(11):32-41. |
| Cui S, Wang ZP, Yu JH, et al. Event-specific detection methods of genetically modified rice BPL9K-2[J]. China Biotechnology, 2018,38(11):32-41. | |
| [9] |
Rao J, Yang LT, Guo JC, et al. Development of event-specific qualitative and quantitative PCR detection methods for the transgenic maize BVLA430101[J]. Eur Food Res Technol, 2016,242(8):1277-1284.
doi: 10.1007/s00217-015-2631-7 URL |
| [10] | 王叶, 谢家建, 黄春蒙, 等. 转cry1Aa基因抗虫棉整合结构解析及转化体特异性检测方法的建立[J]. 棉花学报, 2017,29(4):307-315. |
| Wang Y, Xie JJ, Huang CM, et al. Integrated structure of the modified cry1Aa gene in cotton and its event-specific detection[J]. Cotton Science, 2017,29(4):307-315. | |
| [11] |
Zhang PQ, Xu JY, Zheng QY, et al. Flanking sequence determination and event specific detection of transgenic wheat B72-8-11b strain[J]. Appl Biochem Biotechnol, 2013,169(5):1523-1530.
doi: 10.1007/s12010-012-9989-9 URL |
| [12] |
Zhang MH, Yu YB, Gao XJ, et al. Event-specific quantitative detection of genetically modified wheat B72-8-11 based on the 3' flanking sequence[J]. Eur Food Res Technol, 2015,240(4):775-782.
doi: 10.1007/s00217-014-2383-9 URL |
| [13] | 闫建俊, 白云凤, 左静静, 等. 转基因马铃薯外源基因插入位点分析及检测方法的建立[J]. 分子植物育种, 2020,18(16):5361-5366. |
| Yan JJ, Bai YF, Zuo JJ, et al. Analysis of insertion site of transgenic potato exogenous gene and the establishment of detection method[J]. Molecular Plant Breeding, 2020,18(16):5361-5366. | |
| [14] | http://www.chinasugar.org.cn/i,35,3430,0.html |
| [15] | 黄应昆, 李文凤. 云南“双高甘蔗”病虫害综合防治[J]. 云南农业科技, 2004(4):16-18. |
| Huang YK, Li WF. Comprehensive prevention and control of diseases and pests of Yunnan “double high sugarcane”[J]. Yunnan Agricultural Science and Technology, 2004(4):16-18. | |
| [16] | Xie JJ, Yang J, Luo QW, et al. Survey of sugarcane pests in Xianggui sugarcane area and countermeasures of pest control[J]. Plant Diseases and Pests, 2020,11(1):15-18, 23. |
| [17] | 王关林, 方宏筠. 植物基因工程[M]. 第2版. 北京: 科学出版社, 2002. |
| Wang GL, Fang HJ. Plant genetic engineering[M]. 2nd ed. Beijing: Science Press, 2002. | |
| [18] | 崔学强, 张树珍, 沈林波, 等. 转基因甘蔗植株Southern杂交体系的优化[J]. 生物技术通报, 2015,31(12):105-109. |
| Cui XQ, Zhang SZ, Shen LB, et al. The optimization of southern blot for transgenic sugarcane plants[J]. Biotechnology Bulletin, 2015,31(12):105-109. | |
| [19] | 徐纪明, 胡晗, 毛文轩, 等. 利用重测序技术获取转基因植物T-DNA插入位点[J]. 遗传, 2018,40(8):676-682. |
| Xu JM, Hu H, Mao WX, et al. Identifying T-DNA insertion site(s)of transgenic plants by whole-genome resequencing[J]. Hereditas, 2018,40(8):676-682. | |
| [20] |
Zhang J, Zhang X, Tang H, et al. Allele-defined genome of the autopolyploid sugarcane Saccharum spontaneumL.[J]. Nat Genet, 2018,50:1565-1573.
doi: 10.1038/s41588-018-0237-2 URL |
| [21] | 郭超, 何行健, 邓力华, 等. 转基因水稻 BarKasalath-01事件特异性检测[J]. 分子植物育种, 2017,15(11):4466-4475. |
| Guo C, He XJ, Deng LH, et al. Event-specific detection of genetically modified rice BarKasalath-01[J]. Molecular Plant Breeding, 2017,15(11):4466-4475. | |
| [22] | 仲晓芳, 杨静, 贺红利, 等. 基于基因组重测序分析高油酸转基因大豆事件外源T-DNA旁侧序列及特异性检测[J]. 农业生物技术学报, 2018,26(12):2017-2026. |
| Zhong XF, Yang J, He HL, et al. Analysis of the T-DNA flanking sequences and event-specific PCR detection of high-content Oleic acid transgenic soybean(Glycine max)Based on Genome Re-sequencing[J]. Journal of Agricultural Biotechnology, 2018,26(12):2017-2026. | |
| [23] | Chen S, Shen AJ, Zhou XY, et al. Analysis of the flanking sequence and event-specific detection of transgenic line W-4 of Brassica napus[J]. Agr Sci Tech, 2014,15(7):1089-1094. |
| [24] |
Zhang N, Xu W, Bai W, et al. Event-specific qualitative and quantitative PCR detection of LY038 maize in mixed samples[J]. Food Control, 2011,22(8):1287-1295.
doi: 10.1016/j.foodcont.2011.01.030 URL |
| [25] |
Marmiroli N, Maestri E, Gulli M, et al. Methods for detection of GMOs in food and feed[J]. Anal Bioanal Chem, 2008,392(3):369-384.
doi: 10.1007/s00216-008-2303-6 pmid: 18726090 |
| No related articles found! |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||