








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (1): 33-43.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0793
Previous Articles Next Articles
ZHANG Yuan( ), ZHANG Xue-ping, ZHANG Yue-qian, LI Xiao-juan(
), ZHANG Xue-ping, ZHANG Yue-qian, LI Xiao-juan( )
)
Received:2021-06-24
Online:2022-01-26
Published:2022-02-22
Contact:
LI Xiao-juan
E-mail:zhangyuan@bjfu.edu.cn;lixj@bjfu.edu.cn
ZHANG Yuan, ZHANG Xue-ping, ZHANG Yue-qian, LI Xiao-juan. Advances of Single-molecule Fluorescence Detection Techniques and Applications in Plant Biology[J]. Biotechnology Bulletin, 2022, 38(1): 33-43.
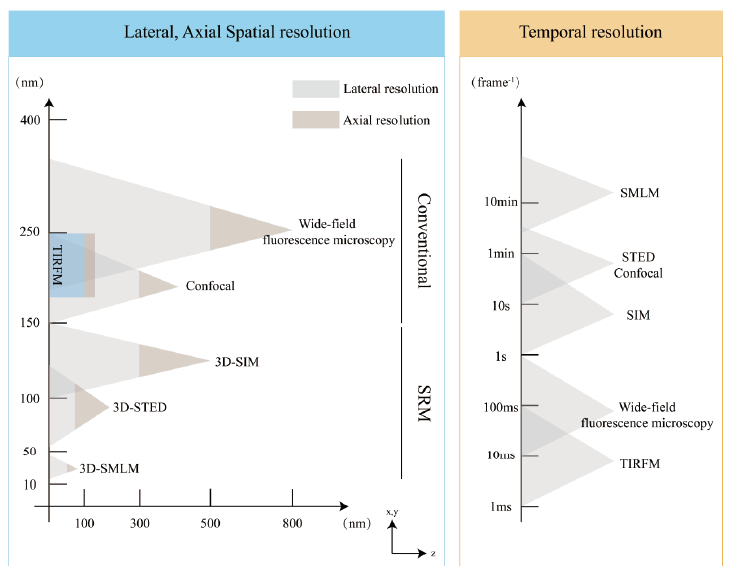
Fig.1 Comparison of spatial and temporal resolution of various imaging techniques The left panel shows spatial resolution of various techniques. The x-axis is the axial resolution,and the y-axis is the lateral resolution of each imaging technique. The right panel shows the time resolution of various techniques. Temporal resolution is estimated based on the lowest exposure times required to image a single plane(SMLM,TIRF)or a volume of a typical mammalian cell(40 µm×40 µm×1 µm)with comparable signal-to-noise-ratio. Refer to Schermelleh et al[46]
| [1] |
Iino R, Iida T, Nakamura A, et al. Single-molecule imaging and manipulation of biomolecular machines and systems[J]. Biochim Biophys Acta Gen Subj, 2018, 1862(2):241-252.
doi: 10.1016/j.bbagen.2017.08.008 URL |
| [2] |
Okamoto K, Hiroshima M, Sako Y. Single-molecule fluorescence-based analysis of protein conformation, interaction, and oligomerization in cellular systems[J]. Biophys Rev, 2018, 10(2):317-326.
doi: 10.1007/s12551-017-0366-3 pmid: 29243093 |
| [3] |
Kapanidis AN, Strick T. Biology, one molecule at a time[J]. Trends Biochem Sci, 2009, 34(5):234-243.
doi: 10.1016/j.tibs.2009.01.008 URL |
| [4] |
Wu WQ, Zhu X, Song CP. Single-molecule technique:a revolutionary approach to exploring fundamental questions in plant science[J]. New Phytol, 2019, 223(2):508-510.
doi: 10.1111/nph.2019.223.issue-2 URL |
| [5] |
Guo AY, Zhang YM, Wang L, et al. Single-molecule imaging in living plant cells:a methodological review[J]. Int J Mol Sci, 2021, 22(10):5071.
doi: 10.3390/ijms22105071 URL |
| [6] |
Taitt CR, Anderson GP, Ligler FS. Evanescent wave fluorescence biosensors[J]. Biosens Bioelectron, 2005, 20(12):2470-2487.
doi: 10.1016/j.bios.2004.10.026 URL |
| [7] |
Liu QL, Chen YH, Liu WJ, et al. Total internal reflection fluorescence pattern-illuminated Fourier ptychographic microscopy[J]. Opt Lasers Eng, 2019, 123:45-52.
doi: 10.1016/j.optlaseng.2019.06.023 URL |
| [8] |
Axelrod D. Cell-substrate contacts illuminated by total internal reflection fluorescence[J]. J Cell Biol, 1981, 89(1):141-145.
doi: 10.1083/jcb.89.1.141 pmid: 7014571 |
| [9] |
Funatsu T, Harada Y, Tokunaga M, et al. Imaging of single fluorescent molecules and individual ATP turnovers by single myosin molecules in aqueous solution[J]. Nature, 1995, 374(6522):555-559.
doi: 10.1038/374555a0 URL |
| [10] |
Mashanov GI, Tacon D, Knight AE, et al. Visualizing single molecules inside living cells using total internal reflection fluorescence microscopy[J]. Methods, 2003, 29(2):142-152.
pmid: 12606220 |
| [11] |
Matsuda K, Sugawa M, Yamagishi M, et al. Visualizing dynamic actin cross-linking processes driven by the actin-binding protein anillin[J]. FEBS Lett, 2020, 594(8):1237-1247.
doi: 10.1002/1873-3468.13720 pmid: 31853940 |
| [12] |
Wan Y, Ash WM, Fan L, et al. Variable-angle total internal reflection fluorescence microscopy of intact cells of Arabidopsis thaliana[J]. Plant Methods, 2011, 7:27.
doi: 10.1186/1746-4811-7-27 URL |
| [13] |
Sun W, Marchuk K, Wang G, et al. Autocalibrated scanning-angle prism-type total internal reflection fluorescence microscopy for nanometer-precision axial position determination[J]. Anal Chem, 2010, 82(6):2441-2447.
doi: 10.1021/ac902789z URL |
| [14] |
Szalai AM, Siarry B, Lukin J, et al. Three-dimensional total-internal reflection fluorescence nanoscopy with nanometric axial resolution by photometric localization of single molecules[J]. Nat Commun, 2021, 12(1):517.
doi: 10.1038/s41467-020-20863-0 URL |
| [15] |
Diekmann R, Helle ØI, Øie CI, et al. Chip-based wide field-of-view nanoscopy[J]. Nat Photonics, 2017, 11(5):322-328.
doi: 10.1038/NPHOTON.2017.55 |
| [16] |
Opstad IS, Ströhl F, Fantham M, et al. A waveguide imaging platform for live-cell TIRF imaging of neurons over large fields of view[J]. bioRxiv, 2019. DOI: 10.1101/2019.12.13.874545.
doi: 10.1101/2019.12.13.874545 |
| [17] |
Feng H, Wang X, Xu Z, et al. Super-resolution fluorescence microscopy for single cell imaging[J]. Adv Exp Med Biol, 2018, 1068:59-71.
doi: 10.1007/978-981-13-0502-3_6 pmid: 29943296 |
| [18] |
Hauser M, Wojcik M, Kim D, et al. Correlative super-resolution microscopy:new dimensions and new opportunities[J]. Chem Rev, 2017, 117(11):7428-7456.
doi: 10.1021/acs.chemrev.6b00604 URL |
| [19] |
Bates M, Huang B, Dempsey GT, et al. Multicolor super-resolution imaging with photo-switchable fluorescent probes[J]. Science, 2007, 317(5845):1749-1753.
pmid: 17702910 |
| [20] |
Flors C, Hotta J, Uji-i H, et al. A stroboscopic approach for fast photoactivation-localization microscopy with Dronpa mutants[J]. J Am Chem Soc, 2007, 129(45):13970-13977.
doi: 10.1021/ja074704l URL |
| [21] |
Geertsema HJ, Aimola G, Fabricius V, et al. Left-handed DNA-PAINT for improved super-resolution imaging in the nucleus[J]. Nat Biotechnol, 2021, 39(5):551-554.
doi: 10.1038/s41587-020-00753-y pmid: 33398151 |
| [22] |
Gu L, Li Y, Zhang S, et al. Molecular resolution imaging by repetitive optical selective exposure[J]. Nat Methods, 2019, 16(11):1114-1118.
doi: 10.1038/s41592-019-0544-2 URL |
| [23] |
Blom H, Widengren J. Stimulated emission depletion microscopy[J]. Chem Rev, 2017, 117(11):7377-7427.
doi: 10.1021/acs.chemrev.6b00653 URL |
| [24] |
Neil MA, Juskaitis R, Wilson T. Method of obtaining optical sectioning by using structured light in a conventional microscope[J]. Opt Lett, 1997, 22(24):1905-1907.
pmid: 18188403 |
| [25] | Guo Y, Li D, Zhang S, et al. Visualizing intracellular organelle and cytoskeletal interactions at nanoscale resolution on millisecond timescales[J]. Cell, 2018, 175(5):1430-1442. e17. |
| [26] |
Huang X, Fan J, Li L, et al. Fast, long-term, super-resolution imaging with Hessian structured illumination microscopy[J]. Nat Biotechnol, 2018, 36(5):451-459.
doi: 10.1038/nbt.4115 URL |
| [27] |
Fritzsche M, Li D, Colin-York H, et al. Self-organizing actin patterns shape membrane architecture but not cell mechanics[J]. Nat Commun, 2017, 8:14347.
doi: 10.1038/ncomms14347 pmid: 28194011 |
| [28] |
Liu B, Xue Y, Zhao W, et al. Three-dimensional super-resolution protein localization correlated with vitrified cellular context[J]. Sci Rep, 2015, 5:13017.
doi: 10.1038/srep13017 URL |
| [29] |
Wojcik M, Hauser M, Li W, et al. Graphene-enabled electron microscopy and correlated super-resolution microscopy of wet cells[J]. Nat Commun, 2015, 6:7384.
doi: 10.1038/ncomms8384 URL |
| [30] |
Pinotsi D, Rodighiero S, Campioni S, et al. An easy path for correlative electron and super-resolution light microscopy[J]. Sci Rep, 2019, 9:15526.
doi: 10.1038/s41598-019-52047-2 URL |
| [31] |
Keren K, Yam PT, Kinkhabwala A, et al. Intracellular fluid flow in rapidly moving cells[J]. Nat Cell Biol, 2009, 11(10):1219-1224.
doi: 10.1038/ncb1965 URL |
| [32] |
Nishizawa K, Fujiwara K, Ikenaga M, et al. Universal glass-forming behavior of in vitro and living cytoplasm[J]. Sci Rep, 2017, 7(1):15143.
doi: 10.1038/s41598-017-14883-y pmid: 29123156 |
| [33] |
Geerts H, De Brabander M, Nuydens R, et al. Nanovid tracking:a new automatic method for the study of mobility in living cells based on colloidal gold and video microscopy[J]. Biophys J, 1987, 52(5):775-782.
pmid: 3427186 |
| [34] |
Shen H, Tauzin LJ, Baiyasi R, et al. Single particle tracking:from theory to biophysical applications[J]. Chem Rev, 2017, 117(11):7331-7376.
doi: 10.1021/acs.chemrev.6b00815 pmid: 28520419 |
| [35] | Kure JL, Andersen CB, Mortensen KI, et al. Revealing plasma membrane nano-domains with diffusion analysis methods[J]. Membranes(Basel), 2020, 10(11):E314. |
| [36] |
Arnspang EC, Sengupta P, Mortensen KI, et al. Regulation of plasma membrane nanodomains of the water channel aquaporin-3 revealed by fixed and live photoactivated localization microscopy[J]. Nano Lett, 2019, 19(2):699-707.
doi: 10.1021/acs.nanolett.8b03721 pmid: 30584808 |
| [37] |
Clausen MP, Arnspang EC, Ballou B, et al. Simultaneous multi-species tracking in live cells with quantum dot conjugates[J]. PLoS One, 2014, 9(6):e97671.
doi: 10.1371/journal.pone.0097671 URL |
| [38] |
Wang W, Shen H, Shuang B, et al. Super temporal-resolved microscopy(STReM)[J]. J Phys Chem Lett, 2016, 7(22):4524-4529.
doi: 10.1021/acs.jpclett.6b02098 URL |
| [39] | Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy(STORM)[J]. Nat Methods, 2006, 3(10):793-795. |
| [40] | Manley S, Gillette JM, Patterson GH, et al. High-density mapping of single-molecule trajectories with photoactivated localization microscopy[J]. Nat Methods, 2008, 5(2):155-157. |
| [41] |
Ram S, Prabhat P, Chao J, et al. High accuracy 3D quantum dot tracking with multifocal plane microscopy for the study of fast intracellular dynamics in live cells[J]. Biophys J, 2008, 95(12):6025-6043.
doi: 10.1529/biophysj.108.140392 URL |
| [42] |
Prabhat P, Ram S, Ward ES, et al. Simultaneous imaging of different focal Planes in fluorescence microscopy for the study of cellular dynamics in three dimensions[J]. IEEE Trans Nanobioscience, 2004, 3(4):237-242.
doi: 10.1109/TNB.2004.837899 URL |
| [43] |
Schnitzbauer J, McGorty R, Huang B. 4Pi fluorescence detection and 3D particle localization with a single objective[J]. Opt Express, 2013, 21(17):19701-19708.
doi: 10.1364/OE.21.019701 pmid: 24105517 |
| [44] | Hou SG, Johnson C, Welsher K. Real-time 3D single particle tracking:towards active feedback single molecule spectroscopy in live cells[J]. Molecules, 2019, 24(15):E2826. |
| [45] |
Shin K, Song YH, Goh Y, et al. Two-dimensional and three-dimensional single particle tracking of upconverting nanoparticles in living cells[J]. Int J Mol Sci, 2019, 20(6). DOI: 10.3390/ijms20061424.
doi: 10.3390/ijms20061424 |
| [46] |
Schermelleh L, Ferrand A, Huser T, et al. Super-resolution microscopy demystified[J]. Nat Cell Biol, 2019, 21(1):72-84.
doi: 10.1038/s41556-018-0251-8 pmid: 30602772 |
| [47] | Giakoumakis NN, Rapsomaniki MA, Lygerou Z. Analysis of protein kinetics using fluorescence recovery after photobleaching(FRAP)[J]. Methods Mol Biol Clifton N J, 2017, 1563:243-267. |
| [48] |
Saito T, Matsunaga D, Matsui TS, et al. Determining the domain-level reaction-diffusion properties of an actin-binding protein transgelin-2 within cells[J]. Exp Cell Res, 2021, 404(1):112619.
doi: 10.1016/j.yexcr.2021.112619 URL |
| [49] |
Seabrooke S, Qiu X, Stewart BA. Nonmuscle Myosin II helps regulate synaptic vesicle mobility at the Drosophila neuromuscular junction[J]. BMC Neurosci, 2010, 11:37.
doi: 10.1186/1471-2202-11-37 pmid: 20233422 |
| [50] |
Ooga M, Funaya S, Hashioka Y, et al. Chd9 mediates highly loosened chromatin structure in growing mouse oocytes[J]. Biochem Biophys Res Commun, 2018, 500(3):583-588.
doi: 10.1016/j.bbrc.2018.04.105 URL |
| [51] |
Hauke S, von Appen A, Quidwai T, et al. Specific protein labeling with caged fluorophores for dual-color imaging and super-resolution microscopy in living cells[J]. Chem Sci, 2017, 8(1):559-566.
doi: 10.1039/C6SC02088G URL |
| [52] |
Köster M, Frahm T, Hauser H. Nucleocytoplasmic shuttling revealed by FRAP and FLIP technologies[J]. Curr Opin Biotechnol, 2005, 16(1):28-34.
doi: 10.1016/j.copbio.2004.11.002 URL |
| [53] |
Liesche J, Gao C, Binczycki P, et al. Direct comparison of leaf plasmodesma structure and function in relation to phloem-loading type[J]. Plant Physiol, 2019, 179(4):1768-1778.
doi: 10.1104/pp.18.01353 URL |
| [54] |
Vartak N, Guenther G, Joly F, et al. Intravital dynamic and correlative imaging of mouse livers reveals diffusion-dominated canalicular and flow-augmented ductular bile flux[J]. Hepatology, 2021, 73(4):1531-1550.
doi: 10.1002/hep.v73.4 URL |
| [55] |
Braeckmans K, Remaut K, Vandenbroucke RE, et al. Line FRAP with the confocal laser scanning microscope for diffusion measurements in small regions of 3-D samples[J]. Biophys J, 2007, 92(6):2172-2183.
pmid: 17208970 |
| [56] |
Maurel D, Banala S, Laroche T, et al. Photoactivatable and photoconvertible fluorescent probes for protein labeling[J]. ACS Chem Biol, 2010, 5(5):507-516.
doi: 10.1021/cb1000229 URL |
| [57] |
Mudumbi KC, Schirmer EC, Yang W. Single-point single-molecule FRAP distinguishes inner and outer nuclear membrane protein distribution[J]. Nat Commun, 2016, 7:12562.
doi: 10.1038/ncomms12562 URL |
| [58] |
Tingey M, Li YC, Yang WD. Protocol for single-molecule fluorescence recovery after photobleaching microscopy to analyze the dynamics and spatial locations of nuclear transmembrane proteins in live cells[J]. STAR Protoc, 2021, 2(2):100490.
doi: 10.1016/j.xpro.2021.100490 URL |
| [59] |
Villarruel C, Dawson SP. Quantification of fluctuations from fluorescence correlation spectroscopy experiments in reaction-diffusion systems[J]. Phys Rev E, 2020, 102(5):052407.
doi: 10.1103/PhysRevE.102.052407 URL |
| [60] |
Schneider F, Hernandez-Varas P, Christoffer Lagerholm B, et al. High photon count rates improve the quality of super-resolution fluorescence fluctuation spectroscopy[J]. J Phys D Appl Phys, 2020, 53(16):164003.
doi: 10.1088/1361-6463/ab6cca pmid: 33191951 |
| [61] |
Kaur G, Costa MW, Nefzger CM, et al. Probing transcription factor diffusion dynamics in the living mammalian embryo with photoactivatable fluorescence correlation spectroscopy[J]. Nat Commun, 2013, 4:1637.
doi: 10.1038/ncomms2657 URL |
| [62] |
Yu L, Lei YZ, Ma Y, et al. A comprehensive review of fluorescence correlation spectroscopy[J]. Front Phys, 2021, 9:644450. DOI: 10.3389/fphy, 2021. 644450.
doi: 10.3389/fphy URL |
| [63] |
Bacia K, Schwille P. Practical guidelines for dual-color fluorescence cross-correlation spectroscopy[J]. Nat Protoc, 2007, 2(11):2842-2856.
doi: 10.1038/nprot.2007.410 URL |
| [64] |
Bi H, Yin Y, Pan B, et al. Scanning single-molecule fluorescence correlation spectroscopy enables kinetics study of DNA hairpin folding with a time window from microseconds to seconds[J]. J Phys Chem Lett, 2016, 7(10):1865-1871.
doi: 10.1021/acs.jpclett.6b00720 URL |
| [65] |
Digman MA, Gratton E. Imaging barriers to diffusion by pair correlation functions[J]. Biophys J, 2009, 97(2):665-673.
doi: 10.1016/j.bpj.2009.04.048 URL |
| [66] |
Schwille P, Meyer-Almes FJ, Rigler R. Dual-color fluorescence cross-correlation spectroscopy for multicomponent diffusional analysis in solution[J]. Biophys J, 1997, 72(4):1878-1886.
pmid: 9083691 |
| [67] |
Zhang X, Cui Y, Yu M, et al. Single-molecule techniques for imaging exo-endocytosis coupling in cells[J]. Trends Plant Sci, 2019, 24(9):879-880.
doi: S1360-1385(19)30143-8 pmid: 31279733 |
| [68] |
Thompson NL, Burghardt TP, Axelrod D. Measuring surface dynamics of biomolecules by total internal reflection fluorescence with photobleaching recovery or correlation spectroscopy[J]. Biophys J, 1981, 33(3):435-454.
pmid: 7225515 |
| [69] |
Hassler K, Leutenegger M, Rigler P, et al. Total internal reflection fluorescence correlation spectroscopy(TIR-FCS)with low background and high count-rate per molecule[J]. Opt Express, 2005, 13(19):7415-7423.
pmid: 19498766 |
| [70] |
Kastrup L, Blom H, Eggeling C, et al. Fluorescence fluctuation spectroscopy in subdiffraction focal volumes[J]. Phys Rev Lett, 2005, 94(17):178104.
pmid: 15904340 |
| [71] |
Eggeling C, Ringemann C, Medda R, et al. Direct observation of the nanoscale dynamics of membrane lipids in a living cell[J]. Nature, 2009, 457(7233):1159-1162.
doi: 10.1038/nature07596 URL |
| [72] |
Honigmann A, Mueller V, Ta H, et al. Scanning STED-FCS reveals spatiotemporal heterogeneity of lipid interaction in the plasma membrane of living cells[J]. Nat Commun, 2014, 5:5412.
doi: 10.1038/ncomms6412 pmid: 25410140 |
| [73] |
Ha T. Single-molecule fluorescence resonance energy transfer[J]. Methods, 2001, 25(1):78-86.
pmid: 11558999 |
| [74] |
Sasmal DK, Pulido LE, Kasal S, et al. Single-molecule fluorescence resonance energy transfer in molecular biology[J]. Nanoscale, 2016, 8(48):19928-19944.
doi: 10.1039/C6NR06794H URL |
| [75] | Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET[J]. Nat Methods, 2008, 5(6):507-516. |
| [76] |
Yu J, Xiao J, Ren X, et al. Probing gene expression in live cells, one protein molecule at a time[J]. Science, 2006, 311(5767):1600-1603.
doi: 10.1126/science.1119623 URL |
| [77] |
Myong S, Rasnik I, Joo C, et al. Repetitive shuttling of a motor protein on DNA[J]. Nature, 2005, 437(7063):1321-1325.
doi: 10.1038/nature04049 URL |
| [78] |
Panda D, Debnath M, Mandal S, et al. A nucleus-imaging probe that selectively stabilizes a minor conformation of c-MYC G-quadruplex and down-regulates c-MYC transcription in human cancer cells[J]. Sci Rep, 2015, 5:13183.
doi: 10.1038/srep13183 URL |
| [79] |
McKinney SA, Joo C, Ha T. Analysis of single-molecule FRET trajectories using hidden Markov modeling[J]. Biophys J, 2006, 91(5):1941-1951.
pmid: 16766620 |
| [80] | Reck-Peterson SL, Derr ND, Stuurman N. Imaging single molecules using total internal reflection fluorescence microscopy(TIRFM)[J]. Cold Spring Harb Protoc, 2010, 2010(3):pdb.top73. |
| [81] |
Zhu P, Craighead HG. Zero-mode waveguides for single-molecule analysis[J]. Annu Rev Biophys, 2012, 41:269-293.
doi: 10.1146/biophys.2012.41.issue-1 URL |
| [82] |
Peng S, Sun R, Wang W, et al. Single-molecule photoactivation FRET:a general and easy-to-implement approach to break the concentration barrier[J]. Angew Chem Int Ed Engl, 2017, 56(24):6882-6885.
doi: 10.1002/anie.201702731 URL |
| [83] |
Jungmann R, Avendaño MS, Woehrstein JB, et al. Multiplexed 3D cellular super-resolution imaging with DNA-PAINT and Exchange-PAINT[J]. Nat Methods, 2014, 11(3):313-318.
doi: 10.1038/nmeth.2835 pmid: 24487583 |
| [84] |
Lee J, Park S, Kang W, et al. Accelerated super-resolution imaging with FRET-PAINT[J]. Mol Brain, 2017, 10(1):63.
doi: 10.1186/s13041-017-0344-5 URL |
| [85] |
Phillips KM, Larson JW, Yantz GR, et al. Application of single molecule technology to rapidly map long DNA and study the conformation of stretched DNA[J]. Nucleic Acids Res, 2005, 33(18):5829-5837.
pmid: 16243782 |
| [86] |
Akkilic N, Geschwindner S, Höök F. Single-molecule biosensors:Recent advances and applications[J]. Biosens Bioelectron, 2020, 151:111944.
doi: S0956-5663(19)31022-X pmid: 31999573 |
| [87] |
Li X, Wang X, Yang Y, et al. Single-molecule analysis of PIP2;1 dynamics and partitioning reveals multiple modes of Arabidopsis plasma membrane aquaporin regulation[J]. Plant Cell, 2011, 23(10):3780-3797.
doi: 10.1105/tpc.111.091454 URL |
| [88] |
Cui Y, Zhao Y, Lu Y, et al. In vivo single-particle tracking of the aquaporin AtPIP2;1 in stomata reveals cell type-specific dynamics[J]. Plant Physiol, 2021, 185(4):1666-1681.
doi: 10.1093/plphys/kiab007 URL |
| [89] |
Hosy E, Martinière A, Choquet D, et al. Super-resolved and dynamic imaging of membrane proteins in plant cells reveal contrasting kinetic profiles and multiple confinement mechanisms[J]. Mol Plant, 2015, 8(2):339-342.
doi: 10.1016/j.molp.2014.10.006 URL |
| [90] |
Bayle V, Fiche JB, Burny C, et al. Single-particle tracking photoactivated localization microscopy of membrane proteins in living plant tissues[J]. Nat Protoc, 2021, 16(3):1600-1628.
doi: 10.1038/s41596-020-00471-4 URL |
| [91] |
Yu M, Liu H, Dong Z, et al. The dynamics and endocytosis of Flot1 protein in response to flg22 in Arabidopsis[J]. J Plant Physiol, 2017, 215:73-84.
doi: 10.1016/j.jplph.2017.05.010 URL |
| [92] |
Wang L, Li H, Lv X, et al. Spatiotemporal dynamics of the BRI1 receptor and its regulation by membrane microdomains in living Arabidopsis cells[J]. Mol Plant, 2015, 8(9):1334-1349.
doi: 10.1016/j.molp.2015.04.005 pmid: 25896454 |
| [93] |
Song S, Wang HJ, Sun MY, et al. Reactive oxygen species-mediated BIN 2 activity revealed by single-molecule analysis[J]. New Phytol, 2019, 223(2):692-704.
doi: 10.1111/nph.15669 pmid: 30597572 |
| [94] |
Clark NM, Hinde E, Winter CM, et al. Tracking transcription factor mobility and interaction in Arabidopsis roots with fluorescence correlation spectroscopy[J]. Elife, 2016, 5:e14770.
doi: 10.7554/eLife.14770 URL |
| [95] |
Long Y, Stahl Y, Weidtkamp-Peters S, et al. In vivo FRET-FLIM reveals cell-type-specific protein interactions in Arabidopsis roots[J]. Nature, 2017, 548(7665):97-102.
doi: 10.1038/nature23317 URL |
| [96] |
Long Y, Stahl Y, Weidtkamp-Peters S, et al. Optimizing FRET-FLIM labeling conditions to detect nuclear protein interactions at native expression levels in living Arabidopsis roots[J]. Front Plant Sci, 2018, 9:639.
doi: 10.3389/fpls.2018.00639 URL |
| [97] |
Langhans M, Meckel T. Single-molecule detection and tracking in plants[J]. Protoplasma, 2014, 251(2):277-291.
doi: 10.1007/s00709-013-0601-0 pmid: 24385216 |
| [98] |
Liu Z, Lavis LD, Betzig E. Imaging live-cell dynamics and structure at the single-molecule level[J]. Mol Cell, 2015, 58(4):644-659.
doi: 10.1016/j.molcel.2015.02.033 URL |
| [99] |
Chang BJ, Perez Meza VD, Stelzer EHK. csiLSFM combines light-sheet fluorescence microscopy and coherent structured illumination for a lateral resolution below 100 nm[J]. PNAS, 2017, 114(19):4869-4874.
doi: 10.1073/pnas.1609278114 URL |
| [1] | SHANG Xiao-yao, ZHOU Ling-fang, YIN Qian-qian, CHAO Yue-hui. Sequencing and Analysis of Full-length Transcriptome from Medicago truncatula [J]. Biotechnology Bulletin, 2021, 37(8): 131-140. |
| [2] | QIAN Hong-ping, CHEN Bo, LIN Jin-xing, CUI Ya-ning. Recent Advances on Dynamic Regulation and Imaging Techniques of RNA Polymerase II [J]. Biotechnology Bulletin, 2021, 37(4): 293-302. |
| [3] | TANG Yong , LIU Xu. Full-length Sequencing of 16S rRNA Gene and Its Analysis Based on the SMRT Sequencing Technology [J]. Biotechnology Bulletin, 2017, 33(8): 34-39. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||