








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (11): 210-219.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0173
Previous Articles Next Articles
ZHAO Yi1( ), LEI Jian-feng2, LIU Min1, HU Zi-yao1, DAI Pei-hong1, LIU Chao1, LI Yue1, LIU Xiao-dong1(
), LEI Jian-feng2, LIU Min1, HU Zi-yao1, DAI Pei-hong1, LIU Chao1, LI Yue1, LIU Xiao-dong1( )
)
Received:2022-02-15
Online:2022-11-26
Published:2022-12-01
Contact:
LIU Xiao-dong
E-mail:277092776@qq.com;xiaodongliu75@aliyun.com
ZHAO Yi, LEI Jian-feng, LIU Min, HU Zi-yao, DAI Pei-hong, LIU Chao, LI Yue, LIU Xiao-dong. Research on the Carrying Capacity of CLCrV-mediated VIGE System[J]. Biotechnology Bulletin, 2022, 38(11): 210-219.
| 引物Primer | 序列Sequence(5'-3') | 用途Application |
|---|---|---|
| GhBsr-k1-sgRNA1F GhBsr-k1-sgRNA1R | GATTGCAGCGTTTCTACCATGGAT AAACATCCATGGTAGAAACGCTGC | 构建GhBsr-k1-sgRNA1 Construction of GhBsr-k1-sgRNA1 |
| GhBsr-k1-sgRNA2F GhBsr-k1-sgRNA2R | GATTGGTGTACTTGGTAAGATCTT AAACAAGATCTTACCAAGTACACC | 构建GhBsr-k1-sgRNA2 Construction of GhBsr-k1-sgRNA2 |
| GhBsr-k1-sgRNA3F GhBsr-k1-sgRNA3R | GATTGCGCTGGCATTACCATGGGC AAACGCCCATGGTAATGCCAGCGC | 构建GhBsr-k1-sgRNA3 Construction of GhBsr-k1-sgRNA3 |
| GhBsr-k1-sgRNA4F GhBsr-k1-sgRNA4R | GATTGTTAATCAGGGGACTGCAGT AAACACTGCAGTCCCCTGATTAAC | 构建GhBsr-k1-sgRNA4 Construction of GhBsr-k1-sgRNA4 |
| GhBsr-k1-sgRNA5F GhBsr-k1-sgRNA5R | GATTGCTGGACCGGATTTAGCAAT AAACATTGCTAAATCCGGTCCAGC | 构建GhBsr-k1-sgRNA5 Construction of GhBsr-k1-sgRNA5 |
| GhBsr-k1-sgRNA6F GhBsr-k1-sgRNA6R | GATTGGTCGTGCCTACGCTGCAGA AAACTCTGCAGCGTAGGCACGACC | 构建GhBsr-k1-sgRNA6 Construction of GhBsr-k1-sgRNA6 |
| M-GhBsr-k1-F1 M-GhBsr-k1-R1 | CCCTTCAGTCTTTTAATGGC ACTCCATTCTGAAACCCAAG | PCR扩增涵盖GhBsr-k1基因A/D亚组靶位点区域 PCR amplification covers the target site region of GhBsr-k1 gene A/D subgroup |
| M-GhBsr-k1-F2 M-GhBsr-k1-R2 | GTCACAGGATGCACGTAAAG AAAGCCACCCAGAACTCAGA | PCR扩增涵盖GhBsr-k1基因A/D亚组靶位点区域 PCR amplification covers the target site region of GhBsr-k1 gene A/D subgroup |
| M-GhBsr-k1-F3 M-GhBsr-k1-R3 | GAAGTGTGTGCTTAAACTTGTC ACTGCTCTGGTAGGATATGG | PCR扩增涵盖GhBsr-k1基因A/D亚组靶位点区域 PCR amplification covers the target site region of GhBsr-k1 gene A/D subgroup |
| M-GhBsr-k1-F4 M-GhBsr-k1-R4 | GTTTAGAATACAATGCAGAAACTTTC GACAAGTTTAAGCACACACTTC | PCR扩增涵盖GhBsr-k1基因A/D亚组靶位点区域 PCR amplification covers the target site region of GhBsr-k1 gene A/D subgroup |
| M-GhBsr-k1-F5 M-GhBsr-k1-R5 | GGATATCTGCAGCTGTGTCT ACTCCATTCTGAAACCCAAG | PCR扩增涵盖GhBsr-k1基因A/D亚组靶位点区域 PCR amplification covers the target site region of GhBsr-k1 gene A/D subgroup |
| M-GhBsr-k1-F6 M-GhBsr-k1-R6 | ACCATAATGCCCGATACTTGC ATAGAAGTGACACCTGAATCATC | PCR扩增涵盖GhBsr-k1基因A/D亚组靶位点区域 PCR amplification covers the target site region of GhBsr-k1 gene A/D subgroup |
| CLCrVA-F CLCrVA-R | GCTGCTTGTATGTTTGGGTG CAAGGGAGAGTAGTTGGCA | CLCrV-A病毒检测 CLCrV-A virus detection |
Table 1 Primer and application
| 引物Primer | 序列Sequence(5'-3') | 用途Application |
|---|---|---|
| GhBsr-k1-sgRNA1F GhBsr-k1-sgRNA1R | GATTGCAGCGTTTCTACCATGGAT AAACATCCATGGTAGAAACGCTGC | 构建GhBsr-k1-sgRNA1 Construction of GhBsr-k1-sgRNA1 |
| GhBsr-k1-sgRNA2F GhBsr-k1-sgRNA2R | GATTGGTGTACTTGGTAAGATCTT AAACAAGATCTTACCAAGTACACC | 构建GhBsr-k1-sgRNA2 Construction of GhBsr-k1-sgRNA2 |
| GhBsr-k1-sgRNA3F GhBsr-k1-sgRNA3R | GATTGCGCTGGCATTACCATGGGC AAACGCCCATGGTAATGCCAGCGC | 构建GhBsr-k1-sgRNA3 Construction of GhBsr-k1-sgRNA3 |
| GhBsr-k1-sgRNA4F GhBsr-k1-sgRNA4R | GATTGTTAATCAGGGGACTGCAGT AAACACTGCAGTCCCCTGATTAAC | 构建GhBsr-k1-sgRNA4 Construction of GhBsr-k1-sgRNA4 |
| GhBsr-k1-sgRNA5F GhBsr-k1-sgRNA5R | GATTGCTGGACCGGATTTAGCAAT AAACATTGCTAAATCCGGTCCAGC | 构建GhBsr-k1-sgRNA5 Construction of GhBsr-k1-sgRNA5 |
| GhBsr-k1-sgRNA6F GhBsr-k1-sgRNA6R | GATTGGTCGTGCCTACGCTGCAGA AAACTCTGCAGCGTAGGCACGACC | 构建GhBsr-k1-sgRNA6 Construction of GhBsr-k1-sgRNA6 |
| M-GhBsr-k1-F1 M-GhBsr-k1-R1 | CCCTTCAGTCTTTTAATGGC ACTCCATTCTGAAACCCAAG | PCR扩增涵盖GhBsr-k1基因A/D亚组靶位点区域 PCR amplification covers the target site region of GhBsr-k1 gene A/D subgroup |
| M-GhBsr-k1-F2 M-GhBsr-k1-R2 | GTCACAGGATGCACGTAAAG AAAGCCACCCAGAACTCAGA | PCR扩增涵盖GhBsr-k1基因A/D亚组靶位点区域 PCR amplification covers the target site region of GhBsr-k1 gene A/D subgroup |
| M-GhBsr-k1-F3 M-GhBsr-k1-R3 | GAAGTGTGTGCTTAAACTTGTC ACTGCTCTGGTAGGATATGG | PCR扩增涵盖GhBsr-k1基因A/D亚组靶位点区域 PCR amplification covers the target site region of GhBsr-k1 gene A/D subgroup |
| M-GhBsr-k1-F4 M-GhBsr-k1-R4 | GTTTAGAATACAATGCAGAAACTTTC GACAAGTTTAAGCACACACTTC | PCR扩增涵盖GhBsr-k1基因A/D亚组靶位点区域 PCR amplification covers the target site region of GhBsr-k1 gene A/D subgroup |
| M-GhBsr-k1-F5 M-GhBsr-k1-R5 | GGATATCTGCAGCTGTGTCT ACTCCATTCTGAAACCCAAG | PCR扩增涵盖GhBsr-k1基因A/D亚组靶位点区域 PCR amplification covers the target site region of GhBsr-k1 gene A/D subgroup |
| M-GhBsr-k1-F6 M-GhBsr-k1-R6 | ACCATAATGCCCGATACTTGC ATAGAAGTGACACCTGAATCATC | PCR扩增涵盖GhBsr-k1基因A/D亚组靶位点区域 PCR amplification covers the target site region of GhBsr-k1 gene A/D subgroup |
| CLCrVA-F CLCrVA-R | GCTGCTTGTATGTTTGGGTG CAAGGGAGAGTAGTTGGCA | CLCrV-A病毒检测 CLCrV-A virus detection |

Fig. 1 Construction of VIGE editing gene vector A:1-6 are B-Zero-AtU6-26∷GhBsr-k1-sgRNA(1-6)cloning vector digestion. B:1-6 are VIGE editing vector CLCrV-AtU6-26∷GhBsr-k1-sgRNA(1-6)digestion. M:Trans2K Plus II DNA marker

Fig. 2 Gene editing vector screening and mutation detection A:WT:PCR/RE assay of negative control genome before and after the Nco I enzyme digestion,1-5:PCR/RE assay of CLCrV-AtU6-26∷GhBsr-k1-sgRNA1 of 10 single plant sample after Nco I enzyme digestion. B:CLCrV-AtU6-26∷GhBsr-k1-sgRNA1 editing vector of the undigested PCR products lacked Nco I sites(due to the presence of a mutation)that were subsequently purified,cloned,and analyzed by sequencing. C:CLCrV-AtU6-26∷GhBsr-k1-sgRNA1 mutation type sequencing peak map. D:WT:PCR/RE assay of negative control genome before and after the Pst I enzyme digestion. 1:PCR/RE assay of CLCrV-AtU6-26∷GhBsr-k1-sgRNA6 of 10 single plant sample after Pst I enzyme digestion. E:CLCrV-AtU6-26∷GhBsr-k1-sgRNA6 editing vector of the undigested PCR products lacked Pst I sites(due to the presence of a mutation)that were subsequently purified,cloned,and analyzed by sequencing. F:CLCrV-AtU6-26∷GhBsr-k1-sgRNA6 mutation type sequencing peak map. “M” indicates the mutation sequence;“+” are denoted with red capital letters;“-” are shown as red dashes;yellow highlighted the differential bases in A/D subgroup of GhBsr-k1 gene. M:Trans2K Plus II DNA marker

Fig. 3 Prediction of secondary structure of sgRNA A:The secondary structure of sgRNA1. B:The secondary structure of sgRNA2. C:The secondary structure of sgRNA3. D:The secondary structure of sgRNA4. E:The secondary structure of sgRNA5. F:The secondary structure of sgRNA6
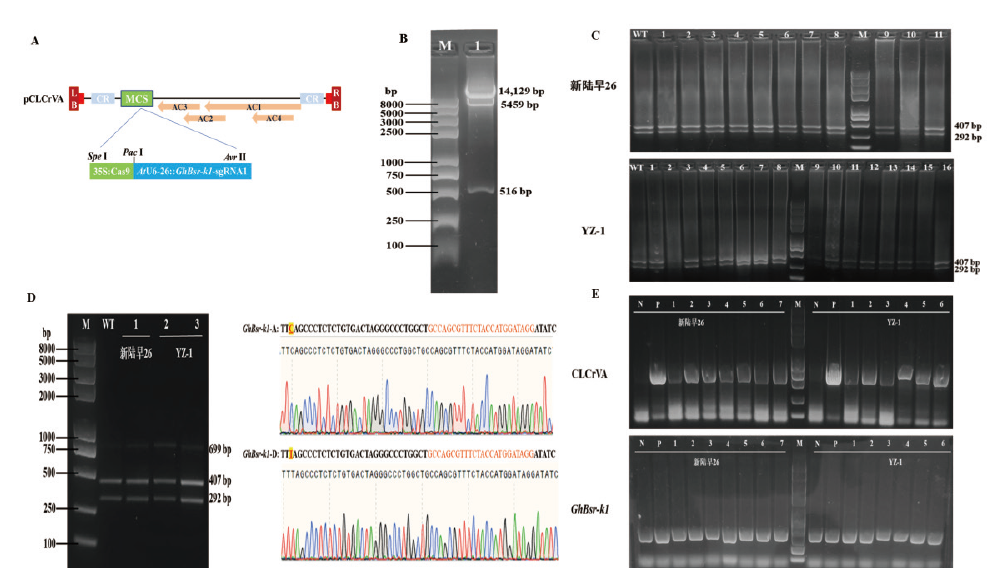
Fig. 4 CLCrV-35S∷Cas9-AtU6-26∷GhBsr-k1-sgRNA1 editing vector construction and mutation detection A:CLCrV-35S∷Cas9-AtU6-26∷GhBsr-k1-sgRNA1 gene editing vector diagram. B:lane 1:CLCrV-35S∷Cas9-AtU6-26∷GhBsr-k1-sgRNA1 gene editing vector was digested with three enzymes. C:PCR/RE assay of CLCrV-35S∷Cas9-AtU6-26∷GhBsr-k1-sgRNA1 of Xinluzao 26 and YZ-1 single plant sample after Nco I enzyme digestion. WT:PCR/RE assay of negative control genome before and after the Nco I enzyme digestion. D:PCR/RE assay of CLCrV-35S∷Cas9-AtU6-26∷GhBsr-k1-sgRNA1 of Xinluzao26 and YZ-1 mix single plant sample after Nco I enzyme digestion. WT:PCR/RE assay of negative control genome before and after the Nco I enzyme digestion.E:Detection of CLCrV-A virus in individual samples of Xinluzao 26 and YZ-1,GhBsr-k1 is internal reference gene. N indicates negative control,and P indicates positive control. M:Trans2K Plus II DNA marker
| [1] |
Gao CX. Genome engineering for crop improvement and future agriculture[J]. Cell, 2021, 184(6):1621-1635.
doi: 10.1016/j.cell.2021.01.005 pmid: 33581057 |
| [2] |
Dupuy P, Sauviac L, Bruand C. Stress-inducible NHEJ in bacteria:function in DNA repair and acquisition of heterologous DNA[J]. Nucleic Acids Res, 2019, 47(3):1335-1349.
doi: 10.1093/nar/gky1212 URL |
| [3] |
Waterworth WM, Drury GE, Bray CM, et al. Repairing breaks in the plant genome:the importance of keeping it together[J]. New Phytol, 2011, 192(4):805-822.
doi: 10.1111/j.1469-8137.2011.03926.x pmid: 21988671 |
| [4] |
Li C, Unver T, Zhang BH. A high-efficiency CRISPR/Cas9 system for targeted mutagenesis in Cotton(Gossypium hirsutum L.)[J]. Sci Rep, 2017, 7:43902.
doi: 10.1038/srep43902 URL |
| [5] | Zhang BH, Rahman MU. Targeted breeding in cotton using CRISPR/Cas 9 genome editing[M]// Rahman M, Zafar Y, Zhang TZ. Cotton Precision Breeding. Cham: Springer International Publishing, 2021:313-327. |
| [6] |
Li C, Zhang BH. Genome editing in cotton using CRISPR/Cas9 system[J]. Methods Mol Biol, 2019, 1902:95-104.
doi: 10.1007/978-1-4939-8952-2_8 pmid: 30543064 |
| [7] |
Juturu VN, Mekala GK, Kirti PB. Current status of tissue culture and genetic transformation research in cotton(Gossypium spp. )[J]. Plant Cell Tiss Organ Cult, 2015, 120(3):813-839.
doi: 10.1007/s11240-014-0640-z URL |
| [8] | Zhang BH, Wang QL, Liu F, et al. Highly efficient plant regeneration through somatic embryogenesis in 20 elite commercial cotton(Gossypium hirsutum L.)cultivars[J]. Plant Omics, 2009, 2(6):259-268. |
| [9] |
Peng RH, Jones DC, Liu F, et al. From sequencing to genome editing for cotton improvement[J]. Trends Biotechnol, 2021, 39(3):221-224.
doi: 10.1016/j.tibtech.2020.09.001 pmid: 32988631 |
| [10] | Oh Y, Kim H, Kim SG. Virus-induced plant genome editing[J]. Curr Opin Plant Biol, 2021, 60:101992. |
| [11] |
Ma XN, Zhang XY, Liu HM, et al. Highly efficient DNA-free plant genome editing using virally delivered CRISPR-Cas9[J]. Nat Plants, 2020, 6(7):773-779.
doi: 10.1038/s41477-020-0704-5 pmid: 32601419 |
| [12] |
Idris AM, Brown JK. Cotton leaf crumple virus is a distinct western hemisphere begomovirus species with complex evolutionary relationships indicative of recombination and reassortment[J]. Phytopathology, 2004, 94(10):1068-1074.
doi: 10.1094/PHYTO.2004.94.10.1068 pmid: 18943795 |
| [13] |
Gu ZH, Huang CJ, Li FF, et al. A versatile system for functional analysis of genes and microRNAs in cotton[J]. Plant Biotechnol J, 2014, 12(5):638-649.
doi: 10.1111/pbi.12169 pmid: 24521483 |
| [14] |
周冠彤, 雷建峰, 代培红, 等. 棉花CRISPR/Cas9基因编辑有效sgRNA高效筛选体系的研究[J]. 作物学报, 2021, 47(3):427-437.
doi: 10.3724/SP.J.1006.2021.04178 |
|
Zhou GT, Lei JF, Dai PH, et al. Efficient screening system of effective sgRNA for cotton CRISPR/Cas9 gene editing[J]. Acta Agron Sin, 2021, 47(3):427-437.
doi: 10.3724/SP.J.1006.2021.04178 URL |
|
| [15] |
Lei JF, Dai PH, Li Y, et al. Heritable gene editing using FT mobile guide RNAs and DNA viruses[J]. Plant Methods, 2021, 17(1):20.
doi: 10.1186/s13007-021-00719-4 pmid: 33596981 |
| [16] |
Zhou XG, Liao HC, Chern M, et al. Loss of function of a rice TPR-domain RNA-binding protein confers broad-spectrum disease resistance[J]. Proc Natl Acad Sci USA, 2018, 115(12):3174-3179.
doi: 10.1073/pnas.1705927115 URL |
| [17] |
Li Y, Zhou YJ, Dai PH, et al. Cotton Bsr-k1 modulates lignin deposition participating in plant resistance against Verticillium dahliae and Fusarium oxysporum[J]. Plant Growth Regul, 2021, 95(2):283-292.
doi: 10.1007/s10725-021-00742-4 URL |
| [18] |
Gao W, Long L, Tian XQ, et al. Genome editing in cotton with the CRISPR/Cas9 system[J]. Front Plant Sci, 2017, 8:1364.
doi: 10.3389/fpls.2017.01364 pmid: 28824692 |
| [19] |
Lorenz R, Bernhart SH, Höner Zu Siederdissen C, et al. ViennaRNA package 2. 0[J]. Algorithms Mol Biol, 2011, 6:26.
doi: 10.1186/1748-7188-6-26 URL |
| [20] |
Liang G, Zhang HM, Lou DJ, et al. Selection of highly efficient sgRNAs for CRISPR/Cas9-based plant genome editing[J]. Sci Rep, 2016, 6:21451.
doi: 10.1038/srep21451 pmid: 26891616 |
| [21] |
Cody WB, Scholthof HB. Plant virus vectors 3. 0:transitioning into synthetic genomics[J]. Annu Rev Phytopathol, 2019, 57:211-230.
doi: 10.1146/annurev-phyto-082718-100301 URL |
| [22] |
Luo YJ, Na R, Nowak JS, et al. Development of a Csy4-processed guide RNA delivery system with soybean-infecting virus ALSV for genome editing[J]. BMC Plant Biol, 2021, 21(1):419.
doi: 10.1186/s12870-021-03138-8 pmid: 34517842 |
| [23] |
Hu JC, Li SY, Li ZL, et al. A barley stripe mosaic virus-based guide RNA delivery system for targeted mutagenesis in wheat and maize[J]. Mol Plant Pathol, 2019, 20(10):1463-1474.
doi: 10.1111/mpp.12849 pmid: 31273916 |
| [24] | 李继洋, 雷建峰, 代培红, 等. 基于棉花U6启动子的海岛棉CRISPR/Cas9基因组编辑体系的建立[J]. 作物学报, 2018, 44(2):227-235. |
|
Li JY, Lei JF, Dai PH, et al. Establishment of CRISPR/Cas9 genome editing system based on GbU6 promoters in cotton(Gossypium barbadense L.)[J]. Acta Agron Sin, 2018, 44(2):227-235.
doi: 10.3724/SP.J.1006.2018.00227 URL |
|
| [25] |
Fu YF, Foden JA, Khayter C, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells[J]. Nat Biotechnol, 2013, 31(9):822-826.
doi: 10.1038/nbt.2623 pmid: 23792628 |
| [26] |
Ma XL, Zhang QY, Zhu QL, et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants[J]. Mol Plant, 2015, 8(8):1274-1284.
doi: 10.1016/j.molp.2015.04.007 pmid: 25917172 |
| [27] |
Ariga H, Toki S, Ishibashi K. Potato virus X vector-mediated DNA-free genome editing in plants[J]. Plant Cell Physiol, 2020, 61(11):1946-1953.
doi: 10.1093/pcp/pcaa123 pmid: 32991731 |
| [28] |
Pausch P, Al-Shayeb B, Bisom-Rapp E, et al. CRISPR-CasΦ from huge phages is a hypercompact genome editor[J]. Science, 2020, 369(6501):333-337.
doi: 10.1126/science.abb1400 pmid: 32675376 |
| [29] |
Wu ZW, Zhang YF, Yu HP, et al. Programmed genome editing by a miniature CRISPR-Cas12f nuclease[J]. Nat Chem Biol, 2021, 17(11):1132-1138.
doi: 10.1038/s41589-021-00868-6 pmid: 34475565 |
| [30] |
Ellison EE, Nagalakshmi U, Gamo ME, et al. Multiplexed heritable gene editing using RNA viruses and mobile single guide RNAs[J]. Nat Plants, 2020, 6(6):620-624.
doi: 10.1038/s41477-020-0670-y pmid: 32483329 |
| [1] | LOU Hui, ZHU Jin-cheng, YANG Yang, ZHANG Wei. Effects of Root Exudates in Resistant and Susceptible Varieties of Cotton on the Growths and Gene Expressions of Fusarium oxysporum [J]. Biotechnology Bulletin, 2023, 39(9): 156-167. |
| [2] | YANG Yang, ZHU Jin-cheng, LOU Hui, HAN Ze-gang, ZHANG Wei. Transcriptome Analysis of Interaction Between Gossypium barbadense and Fusarium oxysporum f. sp. vasinfectum [J]. Biotechnology Bulletin, 2023, 39(6): 259-273. |
| [3] | DENG Jia-hui, LEI Jian-feng, ZHAO Yi, LIU Min, HU Zi-yao, YOU Yang-zi, SHAO Wu-kui, LIU Jian-fei, LIU Xiao-dong. Construction of a New Mini Genome Editing System Based on Csy4 and MCP [J]. Biotechnology Bulletin, 2023, 39(10): 68-79. |
| [4] | ZHU Jin-cheng, YANG Yang, LOU Hui, ZHANG Wei. Regulation of Fusarium wilt Resistance in Cotton by Exogenous Melatonin [J]. Biotechnology Bulletin, 2023, 39(1): 243-252. |
| [5] | LI Xiu-qing, HU Zi-yao, LEI Jian-feng, DAI Pei-hong, LIU Chao, DENG Jia-hui, LIU Min, SUN Ling, LIU Xiao-dong, LI Yue. Cloning and Functional Analysis of Gene GhTIFY9 Related to Cotton Verticillium Wilt Resistance [J]. Biotechnology Bulletin, 2022, 38(8): 127-134. |
| [6] | ZHAO Zeng-qiang, GUO Wen-ting, ZHANG Xi, LI Xiao-ling, ZHANG Wei. Cloning and Functional Analysis of GhERF5-4D Gene Related to Fusarium oxysporum Resistance in Cotton [J]. Biotechnology Bulletin, 2022, 38(4): 193-201. |
| [7] | HU Zi-yao, DAI Pei-hong, LIU Chao, Madina Mulati, WANG Qian, Wugalihan Abuduwili, ZHAO Yi, SUN Ling, XU Shi-jia, LI Yue. Molecular Cloning,Expression and VIGS Construction of a Small GTP-binding Protein Gene GhROP3 in Gossypium hirsutum [J]. Biotechnology Bulletin, 2021, 37(9): 106-113. |
| [8] | LIU Yuan-yuan, YANG Dong-jie, ZUO Dong-yun, CHENG Hai-liang, ZHANG You-ping, LV Li-min, WANG Qiao-lian, SONG Guo-li. Cloning and Functional Verification of GhD6PKL2 from Gossypium hirsutum [J]. Biotechnology Bulletin, 2021, 37(8): 111-120. |
| [9] | WANG Hong-jie, LIU Shao-dong, LIU Rui-hua, ZHANG Si-ping, YANG Jun, PANG Chao-you. Effects of Crop Rotation on Bacterial Communities in Cotton Rhizosphere Soil [J]. Biotechnology Bulletin, 2020, 36(9): 117-124. |
| [10] | SHI Jian-bin, ZHOU Hong, WANG Ning, XU Qing-hua, QIAO Wen-qing, YAN Gen-tu. Purity Identification and Genetic Diversity Analysis of Cotton Germplasm Resources Using SSR Markers [J]. Biotechnology Bulletin, 2018, 34(7): 138-146. |
| [11] | ZHANG Meng-tian, PEI Juan, LI Guo ,ZHAO Hui ,CHEN Jian-quan, ZHU Jian-bo ,WANG Ai-ying. Identification and Pathogenicity Analysis of Cotton Verticillium Wilt from Shihezi Region of Xinjiang [J]. Biotechnology Bulletin, 2018, 34(6): 73-78. |
| [12] | JIANG Lan, PANG Jin-huan, XIAO Wei-lie, ZHANG Guo-li, LIU Jun, YANG Chao. Control Effects of 56 Extracts of Chinese Traditional Medicine on Cotton Wilt Disease [J]. Biotechnology Bulletin, 2018, 34(2): 128-134. |
| [13] | GUO Wen-fang, WANG Nan, LI Gang-qiang, XU Fang-fang, YANG Cai-feng, LIU De-hu. Comparative Analysis of Identification Methods of CP4-EPSPS Transgenic Cotton Plants [J]. Biotechnology Bulletin, 2017, 33(4): 114-118. |
| [14] | XIAO Hai-bing, WANG Peng-jun, LI Xian-feng, DONG Hong-qiang, YANG Ming-lu. Tempo-spatial Distribution of Cry1Ab/c Protein in the Main Stem Leaves of Transgenic Bt Cotton [J]. Biotechnology Bulletin, 2017, 33(12): 108-111. |
| [15] | WANG Shan-shan, WU Hao, LI Liang. Extraction of Volatile Oil from Zanthoxylum and Determination of Its Antibacterial Activity and Insect Resistance [J]. Biotechnology Bulletin, 2017, 33(11): 101-105. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||