








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (9): 156-167.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0284
Previous Articles Next Articles
LOU Hui( ), ZHU Jin-cheng, YANG Yang, ZHANG Wei(
), ZHU Jin-cheng, YANG Yang, ZHANG Wei( )
)
Received:2023-03-29
Online:2023-09-26
Published:2023-10-24
Contact:
ZHANG Wei
E-mail:2930304170@qq.com;zhw_agr@shzu.edu.cn
LOU Hui, ZHU Jin-cheng, YANG Yang, ZHANG Wei. Effects of Root Exudates in Resistant and Susceptible Varieties of Cotton on the Growths and Gene Expressions of Fusarium oxysporum[J]. Biotechnology Bulletin, 2023, 39(9): 156-167.
| 基因名称 Gene name | 引物序列Primer sequence(5'-3') |
|---|---|
| FOTG_00376 | TGCAGCCCTCATCCTTGC(F) |
| ACAGCGCGGATGGTATCG(R) | |
| FOTG_00798 | AGCTGCAATCCCACGCTT(F) |
| AAAAGCAAAGGCAGCGGC(R) | |
| FOTG_00997 | CCAGCGAGTTCTCGACCC(F) |
| CAGAGAAGGGCCGCCAAA(R) | |
| FOTG_01540 | TCTCCAGGGGCCAGTGTT(F) |
| ACTTTGGCCCGATCGCAA(R) | |
| FOTG_02351 | CTTCCCAGGCGTGGCTTT(F) |
| GTCAAACAGGGCGAGCCT(R) | |
| FOTG_05128 | GCCTCATCACCACCCGTC(F) |
| GCGAGACCAGGTGACCAC(R) | |
| FOTG_05209 | AAGACCGGTGCTTCTGCC(F) |
| TGGAGGCGGAGAAGACGA(R) | |
| FOTG_06306 | TCAGCCCTGCCGATGTTG(F) |
| GTCGAGACCGAGCAGCTC(R) | |
| FOTG_07131 | TGGCTCCAAGCTCTGTGC(F) |
| CTGAGTTTCCGTGGCCGT(R) | |
| FOTG_08398 | TGCTTGGTATGGGCGGTG(F) |
| CAGTGCCCAGCTTTCGGA(R) | |
| FOTG_16221 | CCCCAGGAGGCCTCAGAT(F) |
| AGCTCCCGCAAACCTGAC(R) | |
| FOTG_18094 | ACCACCATCCGCAACCTG(F) |
| AGGTCCCGTTCGTTGCTG(R) |
Table 1 Primer sequences
| 基因名称 Gene name | 引物序列Primer sequence(5'-3') |
|---|---|
| FOTG_00376 | TGCAGCCCTCATCCTTGC(F) |
| ACAGCGCGGATGGTATCG(R) | |
| FOTG_00798 | AGCTGCAATCCCACGCTT(F) |
| AAAAGCAAAGGCAGCGGC(R) | |
| FOTG_00997 | CCAGCGAGTTCTCGACCC(F) |
| CAGAGAAGGGCCGCCAAA(R) | |
| FOTG_01540 | TCTCCAGGGGCCAGTGTT(F) |
| ACTTTGGCCCGATCGCAA(R) | |
| FOTG_02351 | CTTCCCAGGCGTGGCTTT(F) |
| GTCAAACAGGGCGAGCCT(R) | |
| FOTG_05128 | GCCTCATCACCACCCGTC(F) |
| GCGAGACCAGGTGACCAC(R) | |
| FOTG_05209 | AAGACCGGTGCTTCTGCC(F) |
| TGGAGGCGGAGAAGACGA(R) | |
| FOTG_06306 | TCAGCCCTGCCGATGTTG(F) |
| GTCGAGACCGAGCAGCTC(R) | |
| FOTG_07131 | TGGCTCCAAGCTCTGTGC(F) |
| CTGAGTTTCCGTGGCCGT(R) | |
| FOTG_08398 | TGCTTGGTATGGGCGGTG(F) |
| CAGTGCCCAGCTTTCGGA(R) | |
| FOTG_16221 | CCCCAGGAGGCCTCAGAT(F) |
| AGCTCCCGCAAACCTGAC(R) | |
| FOTG_18094 | ACCACCATCCGCAACCTG(F) |
| AGGTCCCGTTCGTTGCTG(R) |

Fig. 1 Effects of root exudates on the growth of Fusarium oxysporum A: Effects of root exudates on spore germination. B: Effects of root exudates on F. oxysporum biomass. C and D: Effect of root exudates on bacterial colony diameter. Different letters indicate significant difference P <0.05, the same below
| 样品名称Sample ID | 质控数据Clean reads | 原始数据Raw reads | GC/% | Q20/% | Q30/% | 参考基因组比对Mapped reads |
|---|---|---|---|---|---|---|
| F0a | 23 634 226 | 7 078 961 596 | 52.96 | 98.65 | 95.81 | 44 807 027(94.79%) |
| F0b | 24 801 113 | 7 426 874 552 | 52.97 | 98.43 | 95.31 | 46 773 111(94.30%) |
| G12a | 19 061 986 | 5 703 668 454 | 52.30 | 97.12 | 92.43 | 33 826 843(88.73%) |
| G12b | 22 878 897 | 6 847 717 368 | 52.37 | 98.49 | 95.48 | 41 697 513(91.13%) |
| G24a | 23 252 280 | 6 960 709 832 | 52.42 | 98.34 | 95.12 | 42 523 192(91.44%) |
| G24b | 19 547 876 | 5 853 299 798 | 52.40 | 97.09 | 92.38 | 35 197 149(90.03%) |
| G48a | 28 294 193 | 8 474 124 618 | 52.58 | 98.22 | 94.77 | 51 256 558(90.58%) |
| G48b | 23 044 105 | 6 899 543 008 | 52.51 | 97.29 | 92.79 | 41 817 664(90.73%) |
| G6a | 25 066 610 | 7 505 211 560 | 52.68 | 98.33 | 95.05 | 45 107 683(89.98%) |
| G6b | 22 483 818 | 6 732 866 920 | 52.70 | 98.51 | 95.51 | 40 929 019(91.02%) |
| K12a | 20 781 035 | 6 222 244 284 | 52.26 | 96.85 | 91.94 | 37 148 780(89.38%) |
| K12b | 20 396 915 | 6 107 771 748 | 52.20 | 97.50 | 93.22 | 36 813 504(90.24%) |
| K24a | 22 395 810 | 6 701 578 958 | 52.30 | 97.46 | 93.20 | 39 332 014(87.81%) |
| K24b | 19 324 066 | 5 781 190 570 | 52.20 | 97.49 | 93.24 | 33 512 754(86.71%) |
| K48a | 20 253 195 | 6 063 580 670 | 52.59 | 97.32 | 92.80 | 37 310 755(92.11%) |
| K48b | 23 431 421 | 7 015 769 462 | 52.54 | 98.63 | 95.77 | 42 942 424(91.63%) |
| K6a | 19 328 402 | 5 788 003 554 | 52.75 | 97.24 | 92.64 | 34 813 490(90.06%) |
| K6b | 20 460 546 | 6 123 145 102 | 52.66 | 97.52 | 93.25 | 36 420 714(89.00%) |
| W12a | 20 317 585 | 6 085 191 946 | 52.29 | 98.25 | 94.84 | 37 516 540(92.33%) |
| W12b | 23 399 026 | 7 007 306 032 | 52.23 | 98.25 | 94.86 | 42 862 234(91.59%) |
| W24a | 22 727 561 | 6 805 789 076 | 52.39 | 98.09 | 94.46 | 42 220 799(92.88%) |
| W24b | 21 749 990 | 6 509 670 558 | 52.28 | 98.27 | 94.83 | 40 299 468(92.64%) |
| W48a | 26 212 102 | 7 848 652 478 | 52.73 | 98.42 | 95.30 | 49 390 772(94.21%) |
| W48b | 25 173 938 | 7 534 704 604 | 52.75 | 97.93 | 94.12 | 47 156 136(93.66%) |
| W6a | 22 480 503 | 6 733 786 866 | 52.67 | 98.44 | 95.33 | 41 135 869(91.49%) |
| W6b | 26 838 538 | 8 034 600 906 | 52.81 | 98.48 | 95.43 | 49 740 879(92.67%) |
Table 2 Evaluation statistics of sequencing data
| 样品名称Sample ID | 质控数据Clean reads | 原始数据Raw reads | GC/% | Q20/% | Q30/% | 参考基因组比对Mapped reads |
|---|---|---|---|---|---|---|
| F0a | 23 634 226 | 7 078 961 596 | 52.96 | 98.65 | 95.81 | 44 807 027(94.79%) |
| F0b | 24 801 113 | 7 426 874 552 | 52.97 | 98.43 | 95.31 | 46 773 111(94.30%) |
| G12a | 19 061 986 | 5 703 668 454 | 52.30 | 97.12 | 92.43 | 33 826 843(88.73%) |
| G12b | 22 878 897 | 6 847 717 368 | 52.37 | 98.49 | 95.48 | 41 697 513(91.13%) |
| G24a | 23 252 280 | 6 960 709 832 | 52.42 | 98.34 | 95.12 | 42 523 192(91.44%) |
| G24b | 19 547 876 | 5 853 299 798 | 52.40 | 97.09 | 92.38 | 35 197 149(90.03%) |
| G48a | 28 294 193 | 8 474 124 618 | 52.58 | 98.22 | 94.77 | 51 256 558(90.58%) |
| G48b | 23 044 105 | 6 899 543 008 | 52.51 | 97.29 | 92.79 | 41 817 664(90.73%) |
| G6a | 25 066 610 | 7 505 211 560 | 52.68 | 98.33 | 95.05 | 45 107 683(89.98%) |
| G6b | 22 483 818 | 6 732 866 920 | 52.70 | 98.51 | 95.51 | 40 929 019(91.02%) |
| K12a | 20 781 035 | 6 222 244 284 | 52.26 | 96.85 | 91.94 | 37 148 780(89.38%) |
| K12b | 20 396 915 | 6 107 771 748 | 52.20 | 97.50 | 93.22 | 36 813 504(90.24%) |
| K24a | 22 395 810 | 6 701 578 958 | 52.30 | 97.46 | 93.20 | 39 332 014(87.81%) |
| K24b | 19 324 066 | 5 781 190 570 | 52.20 | 97.49 | 93.24 | 33 512 754(86.71%) |
| K48a | 20 253 195 | 6 063 580 670 | 52.59 | 97.32 | 92.80 | 37 310 755(92.11%) |
| K48b | 23 431 421 | 7 015 769 462 | 52.54 | 98.63 | 95.77 | 42 942 424(91.63%) |
| K6a | 19 328 402 | 5 788 003 554 | 52.75 | 97.24 | 92.64 | 34 813 490(90.06%) |
| K6b | 20 460 546 | 6 123 145 102 | 52.66 | 97.52 | 93.25 | 36 420 714(89.00%) |
| W12a | 20 317 585 | 6 085 191 946 | 52.29 | 98.25 | 94.84 | 37 516 540(92.33%) |
| W12b | 23 399 026 | 7 007 306 032 | 52.23 | 98.25 | 94.86 | 42 862 234(91.59%) |
| W24a | 22 727 561 | 6 805 789 076 | 52.39 | 98.09 | 94.46 | 42 220 799(92.88%) |
| W24b | 21 749 990 | 6 509 670 558 | 52.28 | 98.27 | 94.83 | 40 299 468(92.64%) |
| W48a | 26 212 102 | 7 848 652 478 | 52.73 | 98.42 | 95.30 | 49 390 772(94.21%) |
| W48b | 25 173 938 | 7 534 704 604 | 52.75 | 97.93 | 94.12 | 47 156 136(93.66%) |
| W6a | 22 480 503 | 6 733 786 866 | 52.67 | 98.44 | 95.33 | 41 135 869(91.49%) |
| W6b | 26 838 538 | 8 034 600 906 | 52.81 | 98.48 | 95.43 | 49 740 879(92.67%) |
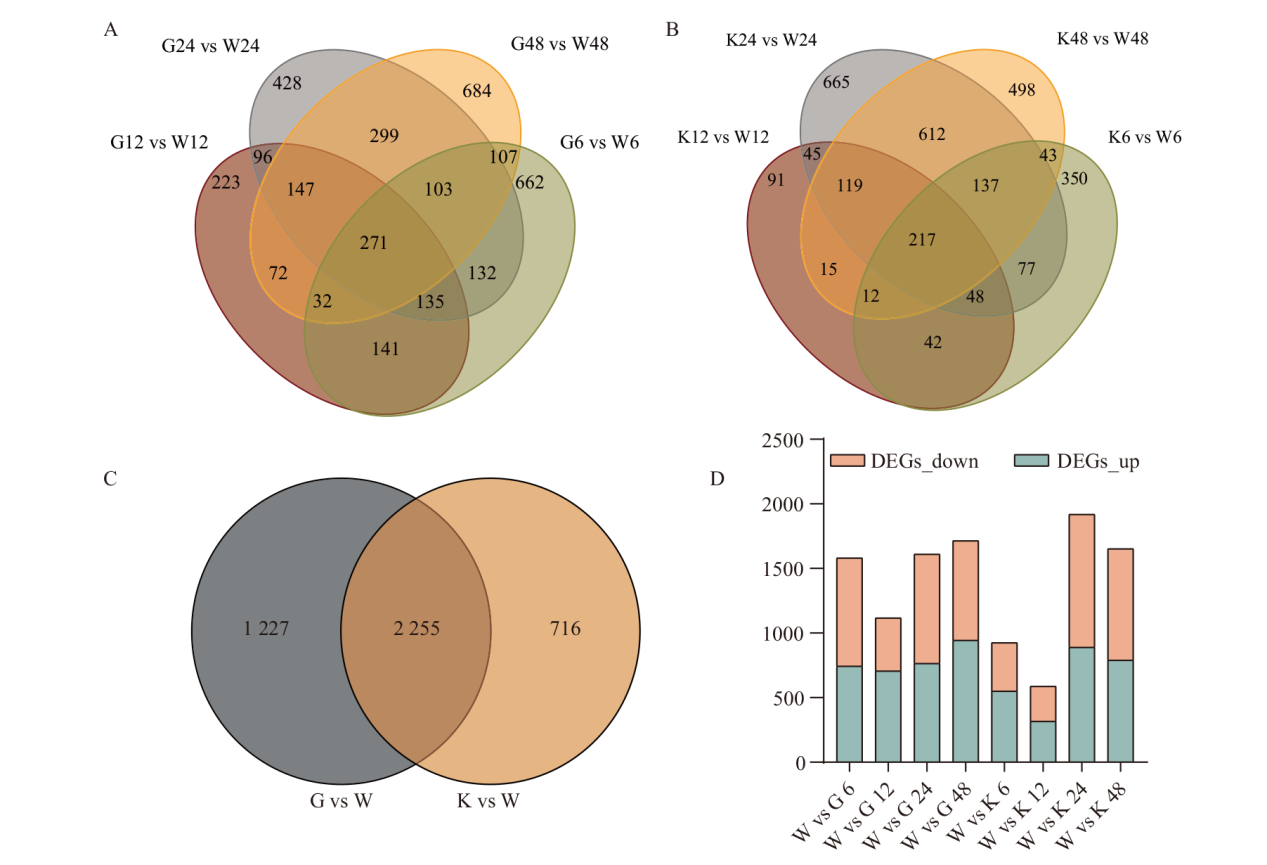
Fig. 4 Differentially expressed genes of F. oxysporum A: Venn diagram of G vs W 6, 12, 24 and 48 h DEGs. B: Venn diagram of G vs W 6, 12, 24 and 48 h DEGs. C: Venn diagram of (G vs W)vs(K vs W). D: K vs W and G vs W 6, 12, 24 and 48 h up and down DEGs
| [1] |
朱金成, 杨洋, 娄慧, 等. 外源褪黑素调控棉花枯萎病抗性研究[J]. 生物技术通报, 2023, 39(1): 243-252.
doi: 10.13560/j.cnki.biotech.bull.1985.2022-0419 |
| Zhu JC, Yang Y, Lou H, et al. Regulation of fusarium wilt resistance in cotton by exogenous melatonin[J]. Biotechnol Bull, 2023, 39(1): 243-252. | |
| [2] |
Zhu Y, Elkins-Arce H, Wheeler TA, et al. Effect of growth stage, cultivar, and root wounding on disease development in cotton caused by Fusarium wilt race 4(Fusarium oxysporum f.sp. vasinfec-tum)[J]. Crop Sci, 2023, 63(1): 101-114.
doi: 10.1002/csc2.v63.1 URL |
| [3] | 王泽华, 方香玲. 尖孢镰刀菌遗传多样性研究进展[J]. 中国草地学报, 2021, 43(5): 106-114. |
| Wang ZH, Fang XL. The research on genetic diversity of Fusarium oxysporum[J]. Chin J Grassland, 2021, 43(5): 106-114. | |
| [4] |
Jia RF, Kang LR, Addrah ME, et al. Potato wilt caused by co-infection of Fusarium spp. and Verticillium dahliae in potato plants[J]. Eur J Plant Pathol, 2023, 165(2): 305-315.
doi: 10.1007/s10658-022-02607-6 |
| [5] |
Yang LH, Zhou Y, Guo LJ, et al. The effect of banana rhizosphere chemotaxis and chemoattractants on Bacillus velezensis LG14-3 root colonization and suppression of banana Fusarium wilt disease[J]. Sustainability, 2022, 15(1): 351.
doi: 10.3390/su15010351 URL |
| [6] |
Li CX, Fu XP, Zhou XG, et al. Treatment with wheat root exudates and soil microorganisms from wheat/watermelon companion cropping can induce watermelon disease resistance against Fusarium ox-ysporum f. sp. niveum[J]. Plant Dis, 2019, 103(7): 1693-1702.
doi: 10.1094/PDIS-08-18-1387-RE URL |
| [7] |
Gai XT, Li S, Jiang N, et al. Comparative transcriptome analysis reveals that ATP synthases regulate Fusarium oxysporum virulence by modulating sugar transporter gene expressions in tobacco[J]. Front Plant Sci, 2022, 13: 978951.
doi: 10.3389/fpls.2022.978951 URL |
| [8] |
Cai HS, Yu N, Liu YY, et al. Meta-analysis of fungal plant pathogen Fusarium oxysporum infection-related gene profiles using transcriptome datasets[J]. Front Microbiol, 2022, 13: 970477.
doi: 10.3389/fmicb.2022.970477 URL |
| [9] |
Lu YX, Dong X, Huang XZ, et al. Combined analysis of the transcriptome and proteome of Eucommia ulmoides Oliv.(Duzhong)in response to Fusarium oxysporum[J]. Front Chem, 2022, 10: 1053227.
doi: 10.3389/fchem.2022.1053227 URL |
| [10] |
Sun YQ, Yang HH, Li JF. Transcriptome analysis reveals the response mechanism of frl-mediated resistance to Fusarium ox-ysporum f. sp. radicis-lycopersici(FORL)infection in tomato[J]. Int J Mol Sci, 2022, 23(13): 7078.
doi: 10.3390/ijms23137078 URL |
| [11] |
Luo Z, Yang XS, Li J, et al. Divergent effects of fertilizer regimes on taxonomic and functional compositions of rhizosphere bacteria and fungi in Phoebe bournei young plantations are associated with root exudates[J]. Forests, 2023, 14(1): 126.
doi: 10.3390/f14010126 URL |
| [12] |
Xu YJ, Chen Z, Li XY, et al. Mycorrhizal fungi alter root exudation to cultivate a beneficial microbiome for plant growth[J]. Funct Ecol, 2023, 37(3): 664-675.
doi: 10.1111/fec.v37.3 URL |
| [13] | 任改弟, 王光飞, 马艳. 根系分泌物与土传病害的关系研究进展[J]. 土壤, 2021, 53(2): 229-235. |
| Ren GD, Wang GF, Ma Y. Research progresses on relationship between plant root exudates and soil-borne diseases[J]. Soils, 2021, 53(2): 229-235. | |
| [14] |
Liang S, Wang YH, Zhang H, et al. Response of root-exuded organic acids in irrigated rice to different water management practices[J]. Eurasian Soil Sci, 2020, 53(11): 1572-1578.
doi: 10.1134/S1064229320110101 |
| [15] |
Chen SC, Ren JJ, Zhao HJ, et al. Trichoderma harzianum improves defense against Fusarium oxysporum by regulating ROS and RNS metabolism, redox balance, and energy flow in cucumber roots[J]. Phytopathology, 2019, 109(6): 972-982.
doi: 10.1094/PHYTO-09-18-0342-R URL |
| [16] |
Lyu JX, Dong Y, Dong K, et al. Intercropping with wheat suppressed Fusarium wilt in faba bean and modulated the composition of root exudates[J]. Plant Soil, 2020, 448(1/2): 153-164.
doi: 10.1007/s11104-019-04413-2 |
| [17] |
Yun TY, Jing T, Zhou DB, et al. Potential biological control of endophytic Streptomyces sp. 5-4 against Fusarium wilt of banana caused by Fusarium oxysporum f. sp. cubense tropical race 4[J]. Phytopathology, 2022, 112(9): 1877-1885.
doi: 10.1094/PHYTO-112-1-0005 URL |
| [18] |
Yang TT, Liu JJ, Li XM, et al. Transcriptomic analysis of Fusarium oxysporum stress-induced pathosystem and screening of fom-2 interaction factors in contrasted melon plants[J]. Front Plant Sci, 2022, 13: 961586.
doi: 10.3389/fpls.2022.961586 URL |
| [19] |
Formela-Luboińska M, Remlein-Starosta D, Waśkiewicz A, et al. The role of saccharides in the mechanisms of pathogenicity of Fusarium oxysporum f. sp. lupini in yellow lupine(Lupinus luteus L.)[J]. Int J Mol Sci, 2020, 21(19): 7258.
doi: 10.3390/ijms21197258 URL |
| [20] |
Li S, Hai J, Wang ZE, et al. Lilium regale Wilson WRKY2 regulates chitinase gene expression during the response to the root rot pathogen Fusarium oxysporum[J]. Front Plant Sci, 2021, 12: 741463.
doi: 10.3389/fpls.2021.741463 URL |
| [21] |
徐丽娇, 郝志鹏, 谢伟, 等. 丛枝菌根真菌根外菌丝跨膜H+和Ca2+流对干旱胁迫的响应[J]. 植物生态学报, 2018, 42(7): 764-773.
doi: 10.17521/cjpe.2018.0089 |
|
Xu LJ, Hao ZP, Xie W, et al. Transmembrane H+ and Ca2+ fluxes through extraradical hyphae of arbuscular mycorrhizal fungi in response to drought stress[J]. Chin J Plant Ecol, 2018, 42(7): 764-773.
doi: 10.17521/cjpe.2018.0089 URL |
|
| [22] |
Berrin JG, Bissaro B. The maize pathogen Ustilago maydis secretes glycoside hydrolases and carbohydrate oxidases directed toward components of the fungal cell wall[J]. Appl Environ Microbiol, 2022, 88(23): e0158122.
doi: 10.1128/aem.01581-22 URL |
| [23] | 侯杨威, 王鑫源, 杨林林, 等. 地黄轮纹病病原菌的细胞壁降解酶活性测定及致病性分析[J]. 北方园艺, 2022(19): 106-113. |
| Hou YW, Wang XY, Yang LL, et al. Determination of cell wall-degrading enzyme activity and pathogenicity analysis of Rotunda pathogens[J]. North Hortic, 2022(19): 106-113. | |
| [24] |
Bhaskar Rao T, Chopperla R, Prathi NB, et al. A comprehensive gene expression profile of pectin degradation enzymes reveals the molecular events during cell wall degradation and pathogenesis of rice sheath blight pathogen Rhizoctonia solani AG1-IA[J]. J Fungi, 2020, 6(2): 71.
doi: 10.3390/jof6020071 URL |
| [25] |
Jeffress S, Arun-Chinnappa K, Stodart B, et al. Genome mining of the citrus pathogen Elsinoë fawcettii; prediction and prioritisation of candidate effectors, cell wall degrading enzymes and secondary metabolite gene clusters[J]. PLoS One, 2020, 15(5): e0227396.
doi: 10.1371/journal.pone.0227396 URL |
| [26] |
Mandalà G, Ceoloni C, Busato I, et al. Transgene pyramiding in wheat: combination of deoxynivalenol detoxification with inhibition of cell wall degrading enzymes to contrast Fusarium Head Blight and Crown Rot[J]. Plant Sci, 2021, 313: 111059.
doi: 10.1016/j.plantsci.2021.111059 URL |
| [27] |
Tini F, Beccari G, Benfield AH, et al. Role of the XylA gene, encoding a cell wall degrading enzyme, during common wheat, durum wheat and barley colonization by Fusarium graminearum[J]. Fungal Genet Biol, 2020, 136: 103318.
doi: 10.1016/j.fgb.2019.103318 URL |
| [28] |
Escobar-Niño A, Morano Bermejo IM, Carrasco Reinado R, et al. Deciphering the dynamics of signaling cascades and virulence factors of B. cinerea during tomato cell wall degradation[J]. Microorganisms, 2021, 9(9): 1837.
doi: 10.3390/microorganisms9091837 URL |
| [29] |
Wang D, Chen JY, Song J, et al. Cytotoxic function of xylanase VdXyn4 in the plant vascular wilt pathogen Verticillium dahli-ae[J]. Plant Physiol, 2021, 187(1): 409-429.
doi: 10.1093/plphys/kiab274 pmid: 34618145 |
| [1] | WU Yuan-ming, LIN Jia-yi, LIU Yu-xi, LI Dan-ting, ZHANG Zong-qiong, ZHENG Xiao-ming, PANG Hong-bo. Identification of Rice Plant Height-associated QTL Using BSA-seq and RNA-seq [J]. Biotechnology Bulletin, 2023, 39(8): 173-184. |
| [2] | FU Yu, JIA Rui-rui, HE He, WANG Liang-gui, YANG Xiu-lian. Growth Differences Among Grafted Seedlings with Two Rootstocks of Catalpa bungei and Comparative Analysis of Transcriptome [J]. Biotechnology Bulletin, 2023, 39(8): 251-261. |
| [3] | ZHAO Lin-yan, XU Wu-mei, WANG Hao-ji, WANG Kun-yan, WEI Fu-gang, YANG Shao-zhou, GUAN Hui-lin. Effects of Applying Biochar on the Rhizosphere Fungal Community and Survival Rate of Panax notoginseng Under Continuous Cropping [J]. Biotechnology Bulletin, 2023, 39(7): 219-227. |
| [4] | YANG Yang, ZHU Jin-cheng, LOU Hui, HAN Ze-gang, ZHANG Wei. Transcriptome Analysis of Interaction Between Gossypium barbadense and Fusarium oxysporum f. sp. vasinfectum [J]. Biotechnology Bulletin, 2023, 39(6): 259-273. |
| [5] | XIE Yang, XING Yu-meng, ZHOU Guo-yan, LIU Mei-yan, YIN Shan-shan, YAN Li-ying. Transcriptome Analysis of Diploid and Autotetraploid in Cucumber Fruit [J]. Biotechnology Bulletin, 2023, 39(3): 152-162. |
| [6] | SHEN Yun-xin, SHI Zhu-feng, ZHOU Xu-dong, LI Ming-gang, ZHANG Qing, FENG Lu-yao, CHEN Qi-bin, YANG Pei-wen. Isolation, Identification and Bio-activity of Three Bacillus Strains with Biocontrol Function [J]. Biotechnology Bulletin, 2023, 39(3): 267-277. |
| [7] | DENG Jia-hui, LEI Jian-feng, ZHAO Yi, LIU Min, HU Zi-yao, YOU Yang-zi, SHAO Wu-kui, LIU Jian-fei, LIU Xiao-dong. Construction of a New Mini Genome Editing System Based on Csy4 and MCP [J]. Biotechnology Bulletin, 2023, 39(10): 68-79. |
| [8] | ZHU Jin-cheng, YANG Yang, LOU Hui, ZHANG Wei. Regulation of Fusarium wilt Resistance in Cotton by Exogenous Melatonin [J]. Biotechnology Bulletin, 2023, 39(1): 243-252. |
| [9] | SUN Zhuo, WANG Yan, HAN Zhong-ming, WANG Yun-he, ZHAO Shu-jie, YANG Li-min. Isolation, Identification and Biocontrol Potential of Rhizospheric Fungus of Saposhnikovia divaricata [J]. Biotechnology Bulletin, 2023, 39(1): 264-273. |
| [10] | LI Ji-hong, JING Yu-ling, MA Gui-zhen, GUO Rong-jun, LI Shi-dong. Genome Construction of Achromobacter 77 and Its Characteristics on Chemotaxis and Antibiotic Resistance [J]. Biotechnology Bulletin, 2022, 38(9): 136-146. |
| [11] | LI Xiu-qing, HU Zi-yao, LEI Jian-feng, DAI Pei-hong, LIU Chao, DENG Jia-hui, LIU Min, SUN Ling, LIU Xiao-dong, LI Yue. Cloning and Functional Analysis of Gene GhTIFY9 Related to Cotton Verticillium Wilt Resistance [J]. Biotechnology Bulletin, 2022, 38(8): 127-134. |
| [12] | ZHAO Zeng-qiang, GUO Wen-ting, ZHANG Xi, LI Xiao-ling, ZHANG Wei. Cloning and Functional Analysis of GhERF5-4D Gene Related to Fusarium oxysporum Resistance in Cotton [J]. Biotechnology Bulletin, 2022, 38(4): 193-201. |
| [13] | ZHAO Yi, LEI Jian-feng, LIU Min, HU Zi-yao, DAI Pei-hong, LIU Chao, LI Yue, LIU Xiao-dong. Research on the Carrying Capacity of CLCrV-mediated VIGE System [J]. Biotechnology Bulletin, 2022, 38(11): 210-219. |
| [14] | HU Zi-yao, DAI Pei-hong, LIU Chao, Madina Mulati, WANG Qian, Wugalihan Abuduwili, ZHAO Yi, SUN Ling, XU Shi-jia, LI Yue. Molecular Cloning,Expression and VIGS Construction of a Small GTP-binding Protein Gene GhROP3 in Gossypium hirsutum [J]. Biotechnology Bulletin, 2021, 37(9): 106-113. |
| [15] | LIU Yuan-yuan, YANG Dong-jie, ZUO Dong-yun, CHENG Hai-liang, ZHANG You-ping, LV Li-min, WANG Qiao-lian, SONG Guo-li. Cloning and Functional Verification of GhD6PKL2 from Gossypium hirsutum [J]. Biotechnology Bulletin, 2021, 37(8): 111-120. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||