








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (12): 287-299.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0347
Previous Articles Next Articles
NIU Hong-yu1( ), SHU Qian2, YANG Hai-jun1, YAN Zhi-yong1, TAN Ju2,3(
), SHU Qian2, YANG Hai-jun1, YAN Zhi-yong1, TAN Ju2,3( )
)
Received:2022-03-22
Online:2022-12-26
Published:2022-12-29
Contact:
TAN Ju
E-mail:nhy@stu.hunau.edu.cn;yhj@hunau.edu.cn
NIU Hong-yu, SHU Qian, YANG Hai-jun, YAN Zhi-yong, TAN Ju. Isolation, Identification, Degradation Characteristics and Metabolic Pathway of an Efficient Sodium Dodecyl Sulfate-degrading Bacterium[J]. Biotechnology Bulletin, 2022, 38(12): 287-299.
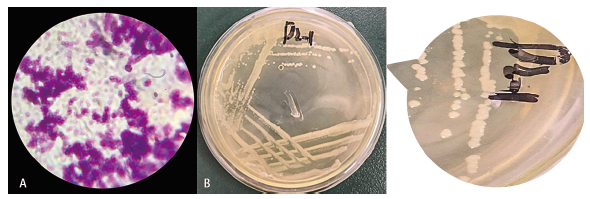
Fig. 3 Colony characteristics of D2 strain A:Gram staining morphology. B:Morphological characteristics of strain in LB solid medium(streaked inoculation)
| 特征 Charateristics | 检测结果 Result |
|---|---|
| 利用柠檬酸盐 Use of citrate | + |
| 糖发酵 Glucose fermentation | + |
| 甲基红 Methyl red | - |
| 尿素水解 Urea hydrolysis | + |
| 产氨 Ammonia | + |
| 苯丙氨酸脱氢酶 Phenylalanine dehydrogenase | - |
| 吲哚 Indocyanine | - |
| 产硫化氢 Hydrogen sulfide | - |
Table 1 Physiological and biochemical characteristics of D2 strain
| 特征 Charateristics | 检测结果 Result |
|---|---|
| 利用柠檬酸盐 Use of citrate | + |
| 糖发酵 Glucose fermentation | + |
| 甲基红 Methyl red | - |
| 尿素水解 Urea hydrolysis | + |
| 产氨 Ammonia | + |
| 苯丙氨酸脱氢酶 Phenylalanine dehydrogenase | - |
| 吲哚 Indocyanine | - |
| 产硫化氢 Hydrogen sulfide | - |
| 物种 Species | NCBI登录号 NCBI accession no. | 相似度 Percent of similarity |
|---|---|---|
| Paraburkholderia tropica strain DSM 15359 | NR_028965.1 | 99.93% |
| Paraburkholderia bannensis strain NBRC 103871 | NR_113178.1 | 98.68% |
| Burkholderia humi Srinivasan strain JCM 18070 | NR_132708.1 | 98.53% |
Table 2 Identification of D2 strain by 16S rDNA gene sequence analysis
| 物种 Species | NCBI登录号 NCBI accession no. | 相似度 Percent of similarity |
|---|---|---|
| Paraburkholderia tropica strain DSM 15359 | NR_028965.1 | 99.93% |
| Paraburkholderia bannensis strain NBRC 103871 | NR_113178.1 | 98.68% |
| Burkholderia humi Srinivasan strain JCM 18070 | NR_132708.1 | 98.53% |

Fig. 5 Effects of environmental factors on the degradation of SDS by strain D2 A:Temperature. B:pH. C:Incubation time. D:NaCl concentration. E:Nitrogen source(1:Sodium nitrate. 2:Ammonium chloride. 3:Sodium nitrate and ammonium chloride. 4:Urea. 5:Peptone). ANOVA Duncan method was used for analysis and different letters means significant difference at P<0.05
| 水平 Level | 因素Factors | ||||
|---|---|---|---|---|---|
| A 温度 Temperature/℃ | B pH | C 时间 Time/h | D 盐度NaCl concentration/% | ||
| 1 | 25 | 6 | 30 | 0.05 | |
| 2 | 30 | 7 | 42 | 0.1 | |
| 3 | 35 | 8 | 48 | 0.3 | |
Table 3 Orthogonal experimental design
| 水平 Level | 因素Factors | ||||
|---|---|---|---|---|---|
| A 温度 Temperature/℃ | B pH | C 时间 Time/h | D 盐度NaCl concentration/% | ||
| 1 | 25 | 6 | 30 | 0.05 | |
| 2 | 30 | 7 | 42 | 0.1 | |
| 3 | 35 | 8 | 48 | 0.3 | |
| 实验号 Experiment No. | 因素 Factors | SDS降解率Degrada-tion rate of SDS/% | |||
|---|---|---|---|---|---|
| A | B | C | D | ||
| 1 | 2 | 1 | 2 | 2 | 81.98 |
| 2 | 2 | 2 | 3 | 1 | 80.29 |
| 3 | 2 | 3 | 1 | 3 | 57.44 |
| 4 | 3 | 3 | 2 | 1 | 83.84 |
| 5 | 1 | 1 | 1 | 1 | 77.13 |
| 6 | 3 | 2 | 1 | 2 | 91.27 |
| 7 | 1 | 3 | 3 | 2 | 49.79 |
| 8 | 1 | 2 | 2 | 3 | 57.30 |
| 9 | 3 | 1 | 3 | 3 | 67.20 |
| K1 | 198.59 | 225.70 | 189.07 | 204.80 | T=646.24 |
| K2 | 253.54 | 233.02 | 215.91 | 224.90 | |
| K3 | 194.12 | 187.52 | 241.27 | 216.55 | |
| X1 | 66.20 | 75.23 | 63.02 | 68.27 | |
| X2 | 84.51 | 77.67 | 71.97 | 74.97 | |
| X3 | 64.71 | 62.51 | 80.42 | 72.18 | |
| R | 19.81 | 15.17 | 17.40 | 6.70 | |
Table 4 Orthogonal experimental results
| 实验号 Experiment No. | 因素 Factors | SDS降解率Degrada-tion rate of SDS/% | |||
|---|---|---|---|---|---|
| A | B | C | D | ||
| 1 | 2 | 1 | 2 | 2 | 81.98 |
| 2 | 2 | 2 | 3 | 1 | 80.29 |
| 3 | 2 | 3 | 1 | 3 | 57.44 |
| 4 | 3 | 3 | 2 | 1 | 83.84 |
| 5 | 1 | 1 | 1 | 1 | 77.13 |
| 6 | 3 | 2 | 1 | 2 | 91.27 |
| 7 | 1 | 3 | 3 | 2 | 49.79 |
| 8 | 1 | 2 | 2 | 3 | 57.30 |
| 9 | 3 | 1 | 3 | 3 | 67.20 |
| K1 | 198.59 | 225.70 | 189.07 | 204.80 | T=646.24 |
| K2 | 253.54 | 233.02 | 215.91 | 224.90 | |
| K3 | 194.12 | 187.52 | 241.27 | 216.55 | |
| X1 | 66.20 | 75.23 | 63.02 | 68.27 | |
| X2 | 84.51 | 77.67 | 71.97 | 74.97 | |
| X3 | 64.71 | 62.51 | 80.42 | 72.18 | |
| R | 19.81 | 15.17 | 17.40 | 6.70 | |
| [1] | Morrison RT, Boyd RN. Organic Chemistry[M]. 4th ed. Massachusetts: Allyn and Bacon Press, 1983. |
| [2] |
Li SP, Su YT, Liu YD, et al. Preparation and characterization of cross-linked enzyme aggregates(CLEAs)of recombinant thermostable alkylsulfatase(SdsAP)from Pseudomonas sp. S9[J]. Process Biochem, 2016, 51(12):2084-2089.
doi: 10.1016/j.procbio.2016.09.013 URL |
| [3] |
Rebello S, Asok AK, Mundayoor S, et al. Surfactants:toxicity, remediation and green surfactants[J]. Environ Chem Lett, 2014, 12(2):275-287.
doi: 10.1007/s10311-014-0466-2 URL |
| [4] |
Lewis MA. Chronic toxicities of surfactants and detergent builders to algae:a review and risk assessment[J]. Ecotoxicol Environ Saf, 1990, 20(2):123-140.
doi: 10.1016/0147-6513(90)90052-7 URL |
| [5] |
Klebensberger J, Rui O, Fritz E, et al. Cell aggregation of Pseudomonas aeruginosa strain PAO1 as an energy-dependent stress response during growth with sodium dodecyl sulfate[J]. Arch Microbiol, 2006, 185(6):417-427.
pmid: 16775748 |
| [6] | Ambily P. Biodegradation of Anionic surfactant Sodium Dodecyl Sulphate(SDS)and analysis of its metabolic products[D]. Meghalaya: Mahatma Gandhi University, 2010 |
| [7] | Agrawal M. Performance of carbonized agricultural waste as a low-cost adsorbent for the removal of sodium dodecyl sulfate from aquatic environment[J]. Int J Sci Eng Res, 2013, 4(6):2909-2913. |
| [8] |
Rahmani Z, Samadi MT. Preparation of magnetic multi-walled carbon nanotubes to adsorb sodium dodecyl sulfate(SDS)[J]. Avicenna J Environ Health Eng, 2017, 4(1):61902.
doi: 10.5812/ajehe.61902 URL |
| [9] | 付凯, 刘志红, 耿超, 等. 改性粉煤灰吸附十二烷基硫酸钠的研究[J]. 矿产保护与利用, 2019, 39(1):90-94, 99. |
| Fu K, Liu ZH, Geng C, et al. Adsorption of sodium dodecyl sulfate by modified fly ash[J]. Conserv Util Miner Resour, 2019, 39(1):90-94, 99. | |
| [10] |
Romanelli MF, Moraes MCF, Villavicencio ALCH, et al. Evaluation of toxicity reduction of sodium dodecyl sulfate submitted to electron beam radiation[J]. Radiat Phys Chem, 2004, 71(1/2):411-413.
doi: 10.1016/j.radphyschem.2004.03.038 URL |
| [11] |
Flilissa A, Méléard P, Darchen A. Selective removal of dodecyl sulfate during electrolysis with aluminum electrodes[J]. Desalination Water Treat, 2013, 51(34/35/36):6719-6728.
doi: 10.1080/19443994.2013.769915 URL |
| [12] |
Aoudjit F, Cherifi O, Halliche D. Simultaneously efficient adsorption and photocatalytic degradation of sodium dodecyl sulfate surfactant by one-pot synthesized TiO2/layered double hydroxide materials[J]. Sep Sci Technol, 2019, 54(7):1095-1105.
doi: 10.1080/01496395.2018.1527352 |
| [13] |
Bhandari PS, Makwana BP, Gogate PR. Microwave and ultrasound assisted dual oxidant based degradation of sodium dodecyl sulfate:efficacy of irradiation approaches and oxidants[J]. J Water Process Eng, 2020, 36:101316.
doi: 10.1016/j.jwpe.2020.101316 URL |
| [14] |
Kıran I, Bektaş N, Cengiz Yatmaz H, et al. Photocatalytic Fenton oxidation of sodium dodecyl sulfate solution using iron-modified zeolite catalyst[J]. Desalination Water Treat, 2013, 51(28/29/30):5768-5775.
doi: 10.1080/19443994.2012.759517 URL |
| [15] |
Mondal B, Adak A, Datta P. Degradation of anionic surfactant in municipal wastewater by UV-H2O2:process optimization using response surface methodology[J]. J Photochem Photobiol A Chem, 2019, 375:237-243.
doi: 10.1016/j.jphotochem.2019.02.030 URL |
| [16] |
Arslan A, Topkaya E, Bingöl D, et al. Removal of anionic surfactant sodium dodecyl sulfate from aqueous solutions by O3/UV/H2O2 advanced oxidation process:process optimization with response surface methodology approach[J]. Sustain Environ Res, 2018, 28(2):65-71.
doi: 10.1016/j.serj.2017.11.002 URL |
| [17] |
Yasin M, Tauseef M, Zafar Z, et al. Plant-microbe synergism in floating treatment wetlands for the enhanced removal of sodium dodecyl sulphate from water[J]. Sustainability, 2021, 13(5):2883.
doi: 10.3390/su13052883 URL |
| [18] |
Othman AR, Yusof MT, Shukor MY. Biodegradation of sodium dodecyl sulphate(SDS)by Serratia marcescens strain DRY6[J]. Bioremed Sci Technol Res, 2019, 7(2):9-14.
doi: 10.54987/bstr.v7i2.486 URL |
| [19] |
Yalaoui-Guellal D, Fella-Temzi S, Djafri-Dib S, et al. The petroleum-degrading bacteria Alcaligenes aquatilis strain YGD 2906 as a potential source of lipopeptide biosurfactant[J]. Fuel, 2021, 285:119112.
doi: 10.1016/j.fuel.2020.119112 URL |
| [20] | Ibrahim AG, Elsalam HEA. Enhancement the biodegradation of sodium dodecyl sulfate by Pseudomonas aeruginosa and Pseudomonas otitidis isolated from waste water in Saudi Arabia[J]. Annu Res Rev Biol, 2018, 28:1-7. |
| [21] |
Zhu Q, Hu ZQ, Ruan MY. Characteristics of sulfate-reducing bacteria and organic bactericides and their potential to mitigate pollution caused by coal gangue acidification[J]. Environ Technol Innov, 2020, 20:101142.
doi: 10.1016/j.eti.2020.101142 URL |
| [22] |
Furmanczyk EM, Kaminski MA, Spolnik G, et al. Isolation and characterization of Pseudomonas spp. strains that efficiently decompose sodium dodecyl sulfate[J]. Front Microbiol, 2017, 8:1872.
doi: 10.3389/fmicb.2017.01872 pmid: 29163375 |
| [23] |
Furmanczyk EM, Kaminski MA, Lipinski L, et al. Pseudomonas laurylsulfatovorans sp. nov., sodium dodecyl sulfate degrading bacteria, isolated from the peaty soil of a wastewater treatment plant[J]. Syst Appl Microbiol, 2018, 41(4):348-354.
doi: S0723-2020(18)30141-3 pmid: 29752019 |
| [24] |
Furmanczyk EM, Lipinski L, Dziembowski A, et al. Genomic and functional characterization of environmental strains of SDS-degrading Pseudomonas spp., providing a source of new sulfatases[J]. Front Microbiol, 2018, 9:1795.
doi: 10.3389/fmicb.2018.01795 pmid: 30174655 |
| [25] |
Halmi MIE, Hussin WSW, Aqlima A, et al. Characterization of a sodium dodecyl sulphate-degrading Pseudomonas sp. strain DRY15 from Antarctic soil[J]. J Environ Biol, 2013, 34(6):1077-1082.
pmid: 24555340 |
| [26] |
Chen YW, Wang L, Dai FZ, et al. Biostimulants application for bacterial metabolic activity promotion and sodium dodecyl sulfate degradation under copper stress[J]. Chemosphere, 2019, 226:736-743.
doi: S0045-6535(19)30626-5 pmid: 30965244 |
| [27] | Panasia G, Philipp B. LaoABCR, a novel system for oxidation of long-chain alcohols derived from SDS and alkane degradation in Pseudomonas aeruginosa[J]. Appl Environ Microbiol, 2018, 84(13):e00626-e00618. |
| [28] |
Hosseini F, Malekzadeh F, Amirmozafari N, et al. Biodegradation of anionic surfactants by isolated bacteria from activated sludge[J]. Int J Environ Sci Technol, 2007, 4(1):127-132.
doi: 10.1007/BF03325970 URL |
| [29] |
Singh KL, Kumar A, Kumar A. Bacillus cereus capable of degrading SDS shows growth with a variety of detergents[J]. World J Microbiol Biotechnol, 1998, 14(5):777-779.
doi: 10.1023/A:1008883915003 URL |
| [30] |
Abboud MM, Khleifat KM, Batarseh M, et al. Different optimization conditions required for enhancing the biodegradation of linear alkylbenzosulfonate and sodium dodecyl sulfate surfactants by novel consortium of Acinetobacter calcoaceticus and Pantoea agglomerans[J]. Enzyme Microb Technol, 2007, 41(4):432-439.
doi: 10.1016/j.enzmictec.2007.03.011 URL |
| [31] | Masdor N, Shukor MSA, Khan A, et al. Isolation and characterization of a molybdenum-reducing and SDS-degrading Klebsiella oxytoca strain Aft-7 and its bioremediation application in the environment[J]. Biodiversitas, 2015, 16(2):238-246. |
| [32] | Adekanmbi AO, Usinola IM. Biodegradation of sodium dodecyl sulphate(SDS)by two bacteria isolated from wastewater generated by a detergent-manufacturing plant in Nigeria[J]. Jordan J Bio Sciences, 2017, 10(4):251-255. |
| [33] |
Chaturvedi V, Kumar A. Metabolism dependent chemotaxis of Pseudomonas aeruginosa N1 towards anionic detergent sodium dodecyl sulfate[J]. Indian J Microbiol, 2014, 54(2):134-138.
doi: 10.1007/s12088-013-0426-8 pmid: 25320412 |
| [34] |
Icgen B, Salik SB, Goksu L, et al. Higher alkyl sulfatase activity required by microbial inhabitants to remove anionic surfactants in the contaminated surface waters[J]. Water Sci Technol, 2017, 76(9/10):2357-2366.
doi: 10.2166/wst.2017.402 URL |
| [35] |
Rusnam M, Gusmanizar N. Characterization of the growth on SDS by Enterobacter sp. strain Neni-13[J]. J Biochem Microbiol Biotechnol, 2017, 5(2):28-32.
doi: 10.54987/jobimb.v5i2.374 URL |
| [36] | Rahman MF, Rusnam M, Gusmanizar N, et al. Molybdate-reducing and SDS-degrading Enterobacter sp. strain neni-13[J]. Nova Biotechnol Chimica, 2016, 15(2):166-181. |
| [37] | 颜丙花, 杨海君, 罗琳, 等. 十二烷基硫酸钠降解菌的分离、鉴定及其降解能力[J]. 化工环保, 2011, 31(2):110-113. |
| Yan BH, Yang HJ, Luo L, et al. Isolation, identification of SDS-degrading strain and its degrading capability[J]. Environ Prot Chem Ind, 2011, 31(2):110-113. | |
| [38] | 刘标, 喻孟元, 王震, 等. 耐镉邻苯二甲酸二辛酯降解菌的筛选及特性研究[J]. 农业资源与环境学报, 2021, 38(2):208-214. |
| Liu B, Yu MY, Wang Z, et al. Isolating and characteristics of a Cd-resistant microorganism used in the biodegradation of di-n-octyl phthalate[J]. J Agric Resour Environ, 2021, 38(2):208-214. | |
| [39] | 李莎, 崔鹤, 尹秀贞, 等. 离子色谱法测定富锂锰基正极材料中的硫酸根[J]. 化学研究与应用, 2021, 33(9):1844-1848. |
| Li S, Cui H, Yin XZ, et al. Determination of sulfate in lithium-riched manganese cathode material by ion chromatography[J]. Chem Res Appl, 2021, 33(9):1844-1848. | |
| [40] | 王发, 徐春燕, 王莉, 等. 毛细管气相色谱法测定聚桂醇原料中乙二醇、月桂醇、二甘醇的含量[J]. 药物分析杂志, 2013, 33(1):138-140. |
| Wang F, Xu CY, Wang L, et al. Capillary GC determination of ethylene glycol, lauryl alcohol and diethylene glycol in lauromacrogol[J]. Chin J Pharm Anal, 2013, 33(1):138-140. | |
| [41] | 王静静, 郭兵, 张庆建, 等. 十通阀双柱切换技术高灵敏检测汽油中甲缩醛和酯类[J]. 分析科学学报, 2019, 35(1):75-79. |
| Wang JJ, Guo B, Zhang QJ, et al. The technology of the ten-pass valve/double column switch for highly sensitive determination of methyl acetals and esters in gasoline[J]. J Anal Sci, 2019, 35(1):75-79. | |
| [42] | 杨丽峰. 洗涤剂中十二酸含量的高效液相色谱法测定[J]. 日用化学品科学, 2013, 36(7):21-23. |
| Yang LF. Determination of lauric acid by HPLC[J]. Deterg Cosmet, 2013, 36(7):21-23. | |
| [43] | 王丹, 吕冰, 周爽, 等. 高效液相色谱法检测淀粉及含淀粉食品中6种有机酸[J]. 中国食品添加剂, 2014(2):108-113. |
| Wang D, Lv B, Zhou S, et al. Determination of six organic acids in starchy food by high performance liquid chromatography[J]. China Food Addit, 2014(2):108-113. | |
| [44] |
Shukor MY, Husin WSW, Rahman MFA, et al. Isolation and characterization of an SDS-degrading Klebsiella oxytoca[J]. J Environ Biol, 2009, 30(1):129-134.
pmid: 20112874 |
| [45] |
Ambily PS, Jisha MS. Metabolic profile of sodium dodecyl sulphate(SDS)biodegradation by Pseudomonas aeruginosa(MTCC 10311)[J]. J Environ Biol, 2014, 35(5):827-831.
pmid: 25204054 |
| [46] | 张若木. 十二烷基硫酸钠降解菌株的筛选与特性研究[D]. 成都: 成都理工大学, 2016. |
| Zhang RM. Screening and characteristics research of SDS degrading strain[D]. Chengdu: Chengdu University of Technology, 2016. | |
| [47] |
Marchesi JR, Owen SA, White GF, et al. SDS-degrading bacteria attach to riverine sediment in response to the surfactant or its primary biodegradation product dodecan-1-ol[J]. Microbiol Read Engl, 1994, 140(Pt 11):2999-3006.
doi: 10.1099/13500872-140-11-2999 URL |
| [48] |
John EM, Rebello S, Asok AK, et al. Pseudomonas plecoglossicida S5, a novel nonpathogenic isolate for sodium dodecyl sulfate degradation[J]. Environ Chem Lett, 2015, 13(1):117-123.
doi: 10.1007/s10311-015-0493-7 URL |
| [1] | RAO Zi-huan, XIE Zhi-xiong. Isolation and Identification of a Cellulose-degrading Strain of Olivibacter jilunii and Analysis of Its Degradability [J]. Biotechnology Bulletin, 2023, 39(8): 283-290. |
| [2] | YOU Zi-juan, CHEN Han-lin, DENG Fu-cai. Research Progress in the Extraction and Functional Activities of Bioactive Peptides from Fish Skin [J]. Biotechnology Bulletin, 2023, 39(7): 91-104. |
| [3] | CHE Yong-mei, GUO Yan-ping, LIU Guang-chao, YE Qing, LI Ya-hua, ZHAO Fang-gui, LIU Xin. Isolation and Identification of Bacterial Strain C8 and B4 and Their Halotolerant Growth-promoting Effects and Mechanisms [J]. Biotechnology Bulletin, 2023, 39(5): 276-285. |
| [4] | WANG Feng-ting, WANG Yan, SUN Ying, CUI Wen-jing, QIAO Kai-bin, PAN Hong-yu, LIU Jin-liang. Isolation and Identification of Saline-alkali Tolerant Aspergillus terreus SYAT-1 and Its Activities Against Plant Pathogenic Fungi [J]. Biotechnology Bulletin, 2023, 39(2): 203-210. |
| [5] | WANG Ya-jun, SI Yun-mei. Screening and Degradation Characteristics of a CP-7 Strain of Dephosphorization Bacteria [J]. Biotechnology Bulletin, 2022, 38(7): 258-268. |
| [6] | ZU Xue, ZHOU Hu, ZHU Hua-jun, REN Zuo-hua, LIU Er-ming. Isolation and Identification of Bacillus subtilis K-268 and Its Biological Control Effect on Rice Blast [J]. Biotechnology Bulletin, 2022, 38(6): 136-146. |
| [7] | WANG Chun-yan, LA Gui-xiao, SU Xiu-hong, LI Meng, DONG Cheng-ming. Screening of Endophytic Bacteria from Rehmannia glutinosa at Different Stages and Analysis of Their Growth-promoting Characteristics [J]. Biotechnology Bulletin, 2022, 38(4): 242-252. |
| [8] | MA Qing-yun, JIANG Xu, LI Qing-qing, SONG Jin-long, ZHOU Yi-qing, RUAN Zhi-yong. Isolation and Identification of Nicosulfuron Degrading Strain Chryseobacterium sp. LAM-M5 and Study on Its Degradation Pathway [J]. Biotechnology Bulletin, 2022, 38(2): 113-122. |
| [9] | ZHANG Gong-you, WANG Yi-han, GUO Min, ZHANG Ting-ting, WANG Bing, LIU Hong-mei. Isolation and Identification of a Cellulase-producing Endophytic Fungus in Paris polyphylla var. yunnanensis [J]. Biotechnology Bulletin, 2022, 38(2): 95-104. |
| [10] | CUI Xin-yu, LI Rong-rong, CAI Rui, WANG Yan, ZHENG Meng-hu, XU Chun-cheng. Isolation,Identification of Lactic Acid Degrading Bacteria in Alfalfa Silage and Their Degradation Characterization [J]. Biotechnology Bulletin, 2021, 37(9): 58-67. |
| [11] | WANG Ying, CHEN Yong-jing, SUN Qing-ye, YANG Meng-yao, WU Dun. Physiological and Biochemical Characteristics of Inquilinus sp. P6-4 Strain and Its Degradation Characteristics for Naphthalene [J]. Biotechnology Bulletin, 2021, 37(6): 117-126. |
| [12] | SHEN Cong, LIU Shuang, WANG Chun-xia, YAN Xue-mei, DAI Jin-xia. Screening,Identification and Characteristics of Petroleum Degrading Bacteria from the Contaminated Soil of Yanchi Aera [J]. Biotechnology Bulletin, 2021, 37(6): 127-135. |
| [13] | XIAO Xiao-shuang, AN Xue-jiao, YE Han-yuan, WANG Lin-ping, ZHONG Bin, ZHANG Qing-hua. Research Progress on Microbial Degradation of Thiocyanate in Wastewater [J]. Biotechnology Bulletin, 2021, 37(2): 224-235. |
| [14] | WEI Xiao-bo, HOU Ying, CHENG Hao-jie, QIN Cui-li, NIU Ming-fu, XU Jian-qiang. Isolation,Identification of Phenol-degrading Pseudoxanthomonas sp. BF-6 and Its Degradation Characteristics and Pathway [J]. Biotechnology Bulletin, 2021, 37(10): 72-80. |
| [15] | LIN Jia-ming, GE Hui, LIN Ke-bing, YANG Zhang-wu, ZHOU Chen, WU Jian-shao, WANG Guo-dong, ZHANG Zhe, YANG Qiu-hua, WANG Yi-lei. Isolation,Identification and Antibiotic Sensitivity Analysis of Bacterial Pathogen from Litopenaeus vannamei with Black Gill Disease [J]. Biotechnology Bulletin, 2020, 36(8): 120-128. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||