








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (6): 298-307.doi: 10.13560/j.cnki.biotech.bull.1985.2022-1372
Previous Articles Next Articles
PAN Hu1,2,3( ), ZHOU Zi-qiong4, TIAN Yun2
), ZHOU Zi-qiong4, TIAN Yun2
Received:2022-11-07
Online:2023-06-26
Published:2023-07-07
Contact:
PAN Hu
E-mail:ph2032007@126.com
PAN Hu, ZHOU Zi-qiong, TIAN Yun. Screening Identification and Degradation Characteristics of Three Iprodione-degrading Strains[J]. Biotechnology Bulletin, 2023, 39(6): 298-307.
| 测试项目Test item | Y-20 | Y-29 | Y-32 | 测试项目Test item | Y-20 | Y-29 | Y-32 | |
|---|---|---|---|---|---|---|---|---|
| 邻硝基苯-半乳糖苷2-Nitrophenyl-galactopyranoside | + | + | - | Kohn明胶Kohn gelatin | + | + | - | |
| 精氨酸Arginine | - | - | - | 葡萄糖Glucose | + | + | - | |
| 赖氨酸Lysine | - | - | - | 甘露醇Mannitol | + | + | - | |
| 鸟氨酸Ornithine | - | - | - | 肌醇Inositol | - | - | - | |
| 柠檬酸钠Sodium citrate | + | +w | + | 山梨醇Sorbitol | - | - | - | |
| 硫代硫酸钠Sodium thiosulfate | - | - | - | 鼠李糖Rhamnose monohydrate | + | - | - | |
| 尿素Urea | - | - | - | 蔗糖Sucrose | + | + | - | |
| 色氨酸Tryptophan(TDA) | + | +w | + | 密二糖Disaccharide | + | - | - | |
| 色氨酸Tryptophan(IND) | - | - | - | 苦杏仁苷Amygdalin | + | + | - | |
| 丙酮酸盐Pyruvate | + | + | - | 阿拉伯糖Arabinose | + | - | +w |
Table 1 Partial physiological and biochemical identification results of strain Y-20, Y-29 and Y-32
| 测试项目Test item | Y-20 | Y-29 | Y-32 | 测试项目Test item | Y-20 | Y-29 | Y-32 | |
|---|---|---|---|---|---|---|---|---|
| 邻硝基苯-半乳糖苷2-Nitrophenyl-galactopyranoside | + | + | - | Kohn明胶Kohn gelatin | + | + | - | |
| 精氨酸Arginine | - | - | - | 葡萄糖Glucose | + | + | - | |
| 赖氨酸Lysine | - | - | - | 甘露醇Mannitol | + | + | - | |
| 鸟氨酸Ornithine | - | - | - | 肌醇Inositol | - | - | - | |
| 柠檬酸钠Sodium citrate | + | +w | + | 山梨醇Sorbitol | - | - | - | |
| 硫代硫酸钠Sodium thiosulfate | - | - | - | 鼠李糖Rhamnose monohydrate | + | - | - | |
| 尿素Urea | - | - | - | 蔗糖Sucrose | + | + | - | |
| 色氨酸Tryptophan(TDA) | + | +w | + | 密二糖Disaccharide | + | - | - | |
| 色氨酸Tryptophan(IND) | - | - | - | 苦杏仁苷Amygdalin | + | + | - | |
| 丙酮酸盐Pyruvate | + | + | - | 阿拉伯糖Arabinose | + | - | +w |
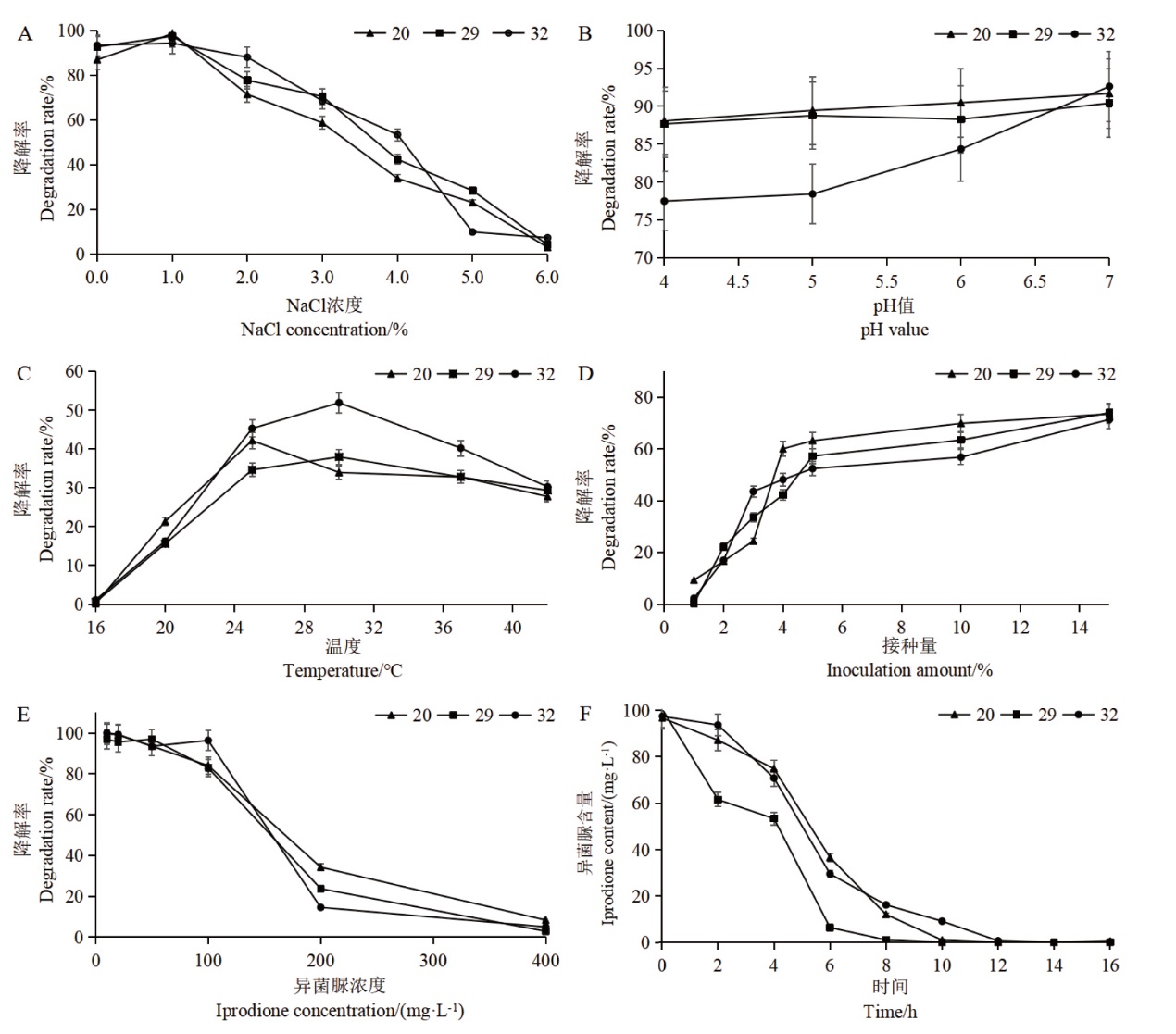
Fig. 4 Effects of NaCl concentration, pH value, temperature, inoculation amount and initial iprodione concentration on the iprodione-degradation rates by strain Y-20, Y-29 and Y-32 and their optimal degradation curves
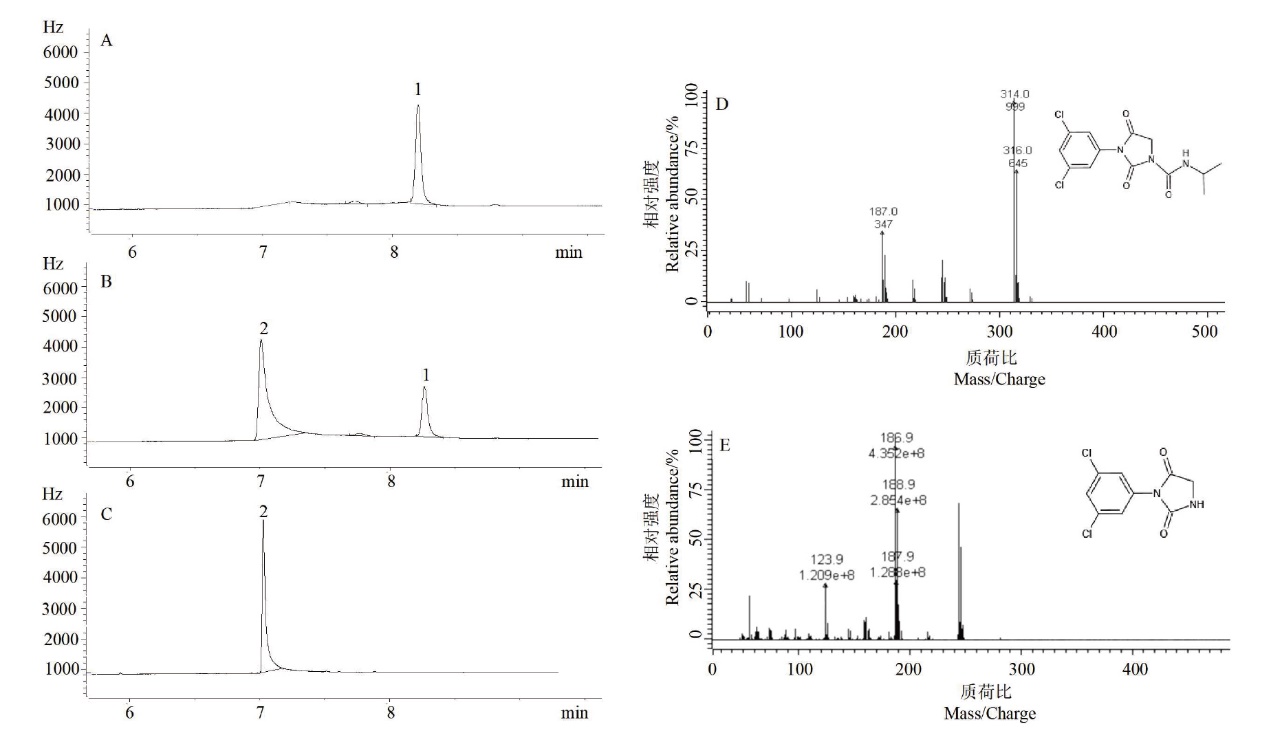
Fig. 5 Change of iprodione and its metabolites in the fermentation liquor and the mass spectrometry analysis results A: Fermentation broth of 0 h. B: Fermentation broth of 6 h. C: Fermentation broth of 12 h. D: Primary mass spectrometry result of compound 1. E: Primary mass spectrometry result of compound 2
| 样品来源 Source | 分离菌株种类 Species of isolates | 异菌脲初始浓度 Initial concentration of iprodione | 降解率 Degradation rate/% | 降解时间 Degradation time | 降解基因 Degradation genes |
|---|---|---|---|---|---|
| 土壤 Soil | Arthrobacter sp. MA6[ | 9.90 mg/L | 86.7 | 7 d | ND |
| 土壤 Soil | Pseudomonas fluorescens, Pseudomonas sp., Pseudomonas paucimobilis[ | 8.25 mg/L | 100 | 20-24 h | ND |
| 土壤 Soil | Zygosaccharomyces rouxii DBVPG 6399[ | 1 mg/L | 100 | 9 d | ND |
| 农田土壤 Farmland soil | Arthrobacter sp.CQH[ | 100 mg/L | 100 | 112 h | ND |
| 土壤 Soil | Microbacterium sp. CHQ-1[ | 100 mg/L | 100 | 96 h | ND |
| 活性污泥 Activated sludge | Microbacterium sp.YJN-G[ | 100 mg/L | 100 | 24 h | ipaH |
| 酸性土壤 Acidic soil | Arthrobacter sp. C1, Achromobacter sp. C2[ | 60 mmol/L | 100 | 10 d | ND |
| 三七根际土壤 Root-zone soil of Panax notoginseng | Bacillus sp.KMS-1[ | 25 mg/L | 41.4 | 7 d | ND |
| 葡萄园种植土壤 Vineyards grow soil | Paenarthrobacter sp. YJN-5[ Paenarthrobacter sp. YJN-D | 1.5 mmol/L | 95 | 80 h | ipaH, ddaH, duaH |
| 大棚土壤 Greenhouse soil | Microbacterium sp. Y-20、Y-29、Y-32 | 100 mg/L | 100 | 8-12 h | ipaH |
Table 2 Basic characteristics of iprodione-degrading strains
| 样品来源 Source | 分离菌株种类 Species of isolates | 异菌脲初始浓度 Initial concentration of iprodione | 降解率 Degradation rate/% | 降解时间 Degradation time | 降解基因 Degradation genes |
|---|---|---|---|---|---|
| 土壤 Soil | Arthrobacter sp. MA6[ | 9.90 mg/L | 86.7 | 7 d | ND |
| 土壤 Soil | Pseudomonas fluorescens, Pseudomonas sp., Pseudomonas paucimobilis[ | 8.25 mg/L | 100 | 20-24 h | ND |
| 土壤 Soil | Zygosaccharomyces rouxii DBVPG 6399[ | 1 mg/L | 100 | 9 d | ND |
| 农田土壤 Farmland soil | Arthrobacter sp.CQH[ | 100 mg/L | 100 | 112 h | ND |
| 土壤 Soil | Microbacterium sp. CHQ-1[ | 100 mg/L | 100 | 96 h | ND |
| 活性污泥 Activated sludge | Microbacterium sp.YJN-G[ | 100 mg/L | 100 | 24 h | ipaH |
| 酸性土壤 Acidic soil | Arthrobacter sp. C1, Achromobacter sp. C2[ | 60 mmol/L | 100 | 10 d | ND |
| 三七根际土壤 Root-zone soil of Panax notoginseng | Bacillus sp.KMS-1[ | 25 mg/L | 41.4 | 7 d | ND |
| 葡萄园种植土壤 Vineyards grow soil | Paenarthrobacter sp. YJN-5[ Paenarthrobacter sp. YJN-D | 1.5 mmol/L | 95 | 80 h | ipaH, ddaH, duaH |
| 大棚土壤 Greenhouse soil | Microbacterium sp. Y-20、Y-29、Y-32 | 100 mg/L | 100 | 8-12 h | ipaH |
| [1] | 来祺. 二甲酰亚胺类杀菌剂代谢产物-3, 5-二氯苯胺的降解行为与生态毒性效应[D]. 南京: 南京农业大学, 2020. |
| Lai Q. Degradation behavior and ecological toxicity of dicarboximide fungicide metabolite-3, 5-dichloroaniline[D]. Nanjing: Nanjing Agricultural University, 2020. | |
| [2] | 陈夕军, 王艳, 童蕴慧, 等. 灰葡萄孢抗二甲酰亚胺类杀菌剂研究进展[J]. 植物保护, 2009, 35(3): 16-19. |
| Chen XJ, Wang Y, Tong YH, et al. Advances in research of the resistance of Botrytis cinerea to dicarboximides fungicides[J]. Plant Prot, 2009, 35(3): 16-19. | |
| [3] | 肖翔. 核盘菌对二甲酰亚胺类杀菌剂菌核净的抗性机理研究[D]. 武汉: 华中农业大学, 2013. |
| Xiao X. Molecular mechanism of Sclerotinia sclerotiorum to the DCF fungicide dimethachlon[D]. Wuhan: Huazhong Agricultural University, 2013. | |
| [4] | Eevers N, White JC, Vangronsveld J, et al. Bio- and phytoremediation of pesticide-contaminated environments: a review[J]. Adv Bot Res, 2017, 83: 277-318. |
| [5] |
Carneiro LS, Martínez LC, Gonçalves WG, et al. The fungicide iprodione affects midgut cells of non-target honey bee Apis mellifera workers[J]. Ecotoxicol Environ Saf, 2020, 189: 109991.
doi: 10.1016/j.ecoenv.2019.109991 URL |
| [6] |
Bernardes PM, Andrade-Vieira LF, Aragão FB, et al. Toxicological effects of comercial formulations of fungicides based on procymidone and iprodione in seedlings and root tip cells of Allium cepa[J]. Environ Sci Pollut Res Int, 2019, 26(20): 21013-21021.
doi: 10.1007/s11356-019-04636-x |
| [7] |
Washington T, Tchounwou PB. Cytotoxicity and transcriptional activation of stress genes in human liver carcinoma(HepG2)cells exposed to iprodione[J]. Int J Environ Res Public Health, 2004, 1(1): 12-20.
doi: 10.3390/ijerph2004010012 URL |
| [8] | Yang ZG, Jiang WK, Wang XH, et al. An amidase gene, ipaH, is responsible for the initial step in the iprodione degradation pathway of Paenarthrobacter sp. strain YJN-5[J]. Appl Environ Microbiol, 2018, 84(19): e01150-e01118. |
| [9] |
Verdenelli RA, Lamarque AL, Meriles JM. Short-term effects of combined iprodione and vermicompost applications on soil microbial community structure[J]. Sci Total Environ, 2012, 414: 210-219.
doi: 10.1016/j.scitotenv.2011.10.066 URL |
| [10] | 汪震, 华修德, 施海燕, 等. 异菌脲在油菜植株、土壤和水中的代谢途径及代谢物3, 5-DCA的毒性研究[J]. 生态毒理学报, 2022, 17(4): 378-385. |
| Wang Z, Hua XD, Shi HY, et al. Studies on metabolic pathways of iprodione in rape plant, soil, and water and toxicity of metabolite 3,5-DCA[J]. Asian J Ecotoxicol, 2022, 17(4): 378-385. | |
| [11] | 杨正中. 异菌脲降解菌的分离鉴定、降解特性及降解途径研究[D]. 南京: 南京农业大学, 2016. |
| Yang ZZ. Isolation & characterization of an iprodione-degrading strain and its degradation pathway[D]. Nanjing: Nanjing Agricultural University, 2016. | |
| [12] |
Cao L, Shi WH, Shu RD, et al. Isolation and characterization of a bacterium able to degrade high concentrations of iprodione[J]. Can J Microbiol, 2018, 64(1): 49-56.
doi: 10.1139/cjm-2017-0185 pmid: 29219613 |
| [13] |
Athiel P, Alfizar, Mercadier C, et al. Degradation of iprodione by a soil Arthrobacter-like strain[J]. Appl Environ Microbiol, 1995, 61(9): 3216-3220.
doi: 10.1128/aem.61.9.3216-3220.1995 URL |
| [14] |
Mercadier C, Vega D, Bastide J, et al. Iprodione degradation by isolated soil microorganisms[J]. FEMS Microbiol Ecol, 1997, 23(3): 207-215.
doi: 10.1111/j.1574-6941.1997.tb00403.x URL |
| [15] | 曹礼, 舒润东, 石文红, 等. 一株降解异菌脲菌株的鉴定及响应面分析法优化其培养条件[J]. 食品工业科技, 2017, 38(6): 168-173, 178. |
| Cao L, Shu RD, Shi WH, et al. Identification of an iprodione-degrading strain and optimization of the culture condition by response surface analysis method[J]. Sci Technol Food Ind, 2017, 38(6): 168-173, 178. | |
| [16] |
Campos M, Karas PS, Perruchon C, et al. Novel insights into the metabolic pathway of iprodione by soil bacteria[J]. Environ Sci Pollut Res Int, 2017, 24(1): 152-163.
doi: 10.1007/s11356-016-7682-1 URL |
| [17] | 杨正中, 吴广, 金文, 等. 异菌脲降解菌YJN-G的分离、鉴定及降解特性[J]. 应用与环境生物学报, 2017, 23(1): 164-168. |
| Yang ZZ, Wu G, Jin W, et al. Isolation, identification and characterization of an iprodione-degrading strain YJN-G[J]. Chin J Appl Environ Biol, 2017, 23(1): 164-168. | |
| [18] | 李艳. 三七种植区土壤农药残留特征及微生物修复研究[D]. 贵阳: 贵州大学, 2018. |
| Li Y. Characteristics of pesticide residues and microbial remediation in soil of Panax notoginseng growing areas[D]. Guiyang: Guizhou University, 2018. | |
| [19] |
Pan H, Zhou J, Dawa ZM, et al. Diversity of culturable bacteria isolated from highland barley cultivation soil in Qamdo, Tibet Autonomous Region[J]. Pol J Microbiol, 2021, 70(1): 87-97.
doi: 10.33073/pjm-2021-008 pmid: 33815530 |
| [20] |
Yoon SH, Ha SM, Kwon S, et al. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies[J]. Int J Syst Evol Microbiol, 2017, 67(5): 1613-1617.
doi: 10.1099/ijsem.0.001755 URL |
| [21] |
Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets[J]. Mol Biol Evol, 2016, 33(7): 1870-1874.
doi: 10.1093/molbev/msw054 pmid: 27004904 |
| [22] | 潘虎. 气相色谱-串联质谱法测定蔬菜、土壤和水中异菌脲及其代谢产物的残留量[J]. 西藏农业科技, 2022, 44(1): 60-65. |
| Pan H. Determination of iprodione and its metabolites residues in vegetables, soil and water by gas chromatography-tandem mass spectrometry[J]. Tibet J Agric Sci, 2022, 44(1): 60-65. | |
| [23] | 刘茜, 张宪, 陈敏, 等. 嘧菌环胺·异菌脲可湿性粉剂在葡萄和土壤中的残留消解动态及风险评估[J]. 农产品质量与安全, 2021(2): 30-35, 41. |
| Liu Q, Zhang X, Chen M, et al. Residue dissipation and dietary risk assessment of cyprodinil and iprodione wettable powder in grape and soil[J]. Qual Saf Agro Prod, 2021(2): 30-35, 41. | |
| [24] |
Zadra C, Cardinali G, Corte L, et al. Biodegradation of the fungicide iprodione by Zygosaccharomyces rouxii strain DBVPG 6399[J]. J Agric Food Chem, 2006, 54(13): 4734-4739.
doi: 10.1021/jf060764j URL |
| [25] |
Zhang ML, Ren YJ, Jiang WK, et al. Comparative genomic analysis of iprodione-degrading Paenarthrobacter strains reveals the iprodione catabolic molecular mechanism in Paenarthrobacter sp. strain YJN-5[J]. Environ Microbiol, 2021, 23(2): 1079-1095.
doi: 10.1111/emi.v23.2 URL |
| [26] | 袁浩亮, 龙泽东, 孙梅, 等. 国际土壤酸化研究知识图谱分析[J]. 土壤通报, 2022, 53(4): 989-997. |
| Yuan HL, Long ZD, Sun M, et al. Knowledge map of international research on soil acidification[J]. Chin J Soil Sci, 2022, 53(4): 989-997. |
| [1] | WU Qiao-yin, SHI You-zhi, LI Lin-lin, PENG Zheng, TAN Zai-yu, LIU Li-ping, ZHANG Juan, PAN Yong. In Situ Screening of Carotenoid Degrading Strains and the Application in Improving Quality and Aroma of Cigar [J]. Biotechnology Bulletin, 2023, 39(9): 192-201. |
| [2] | WANG Yu, YIN Ming-shen, YIN Xiao-yan, XI Jia-qin, YANG Jian-wei, NIU Qiu-hong. Screening, Identification and Degradation Characteristics of Nicotine-degrading Bacteria in Lasioderma serricorne [J]. Biotechnology Bulletin, 2023, 39(6): 308-315. |
| [3] | ZHANG Jing, ZHANG Hao-rui, CAO Yun, HUANG Hong-ying, QU Ping, ZHANG Zhi-ping. Research Progress in Thermophilic Microorganisms for Cellulose Degradation [J]. Biotechnology Bulletin, 2023, 39(6): 73-87. |
| [4] | MA Yu-qian, SUN Dong-hui, YUE Hao-feng, XIN Jia-yu, LIU Ning, CAO Zhi-yan. Identification, Heterologous Expression and Functional Analysis of a GH61 Family Glycoside Hydrolase from Setosphaeria turcica with the Assisting Function in Degrading Cellulose [J]. Biotechnology Bulletin, 2023, 39(4): 124-135. |
| [5] | YANG Dong, TANG Ying. Enzymatic Characterization and Degradation Sites of AFB1 Degradation by the Extracellular Enzyme of Bacillus subtilis Strain WTX1 [J]. Biotechnology Bulletin, 2023, 39(4): 93-102. |
| [6] | ZHANG Ao-jie, LI Qing-yun, SONG Wen-hong, YAN Shao-hui, TANG Ai-xing, LIU You-yan. Whole Genome Sequencing Analysis of a Phenol-degrading Strain Alcaligenes faecalis JF101 [J]. Biotechnology Bulletin, 2023, 39(10): 292-303. |
| [7] | XU Yang, DING Hong, ZHANG Guan-chu, GUO Qing, ZHANG Zhi-meng, DAI Liang-xiang. Metabolomics Analysis of Germinating Peanut Seed Under Salt Stress [J]. Biotechnology Bulletin, 2023, 39(1): 199-213. |
| [8] | LI Liu, MU Ying-chun, LIU Lu, ZHANG Hong-yu, XU Jin-hua, YANG Zhen, QIAO Lu, SONG Jin-long. Research Progress on Contamination Control of Fluoroquinolone Antibiotics and Drug Resistance Genes [J]. Biotechnology Bulletin, 2022, 38(9): 84-95. |
| [9] | WANG Ya-jun, SI Yun-mei. Screening and Degradation Characteristics of a CP-7 Strain of Dephosphorization Bacteria [J]. Biotechnology Bulletin, 2022, 38(7): 258-268. |
| [10] | GULJAMAL·Aisa , XING Jun, LI An, ZHANG Rui. Non-targeted Metabolomics Analysis of Benzo(α)pyrene by Microorganisms in Kefir Grains [J]. Biotechnology Bulletin, 2022, 38(5): 123-135. |
| [11] | WANG Xin-guang, TIAN Lei, WANG En-ze, ZHONG Cheng, TIAN Chun-jie. Construction of Microbial Consortium for Efficient Degradation of Corn Straw and Evaluation of Its Degradation Effect [J]. Biotechnology Bulletin, 2022, 38(4): 217-229. |
| [12] | FU Ya-li, PENG Wan-li, LIN Shuang-jun, DENG Zi-xin, LIANG Ru-bing. Gene Cloning and Enzymatic Properties of the Short Chain Dehydrogenase SDR-X1 from Pseudomonas citronellolis SJTE-3 [J]. Biotechnology Bulletin, 2022, 38(3): 121-129. |
| [13] | MA Qing-yun, JIANG Xu, LI Qing-qing, SONG Jin-long, ZHOU Yi-qing, RUAN Zhi-yong. Isolation and Identification of Nicosulfuron Degrading Strain Chryseobacterium sp. LAM-M5 and Study on Its Degradation Pathway [J]. Biotechnology Bulletin, 2022, 38(2): 113-122. |
| [14] | NIU Hong-yu, SHU Qian, YANG Hai-jun, YAN Zhi-yong, TAN Ju. Isolation, Identification, Degradation Characteristics and Metabolic Pathway of an Efficient Sodium Dodecyl Sulfate-degrading Bacterium [J]. Biotechnology Bulletin, 2022, 38(12): 287-299. |
| [15] | CUI Xin-yu, LI Rong-rong, CAI Rui, WANG Yan, ZHENG Meng-hu, XU Chun-cheng. Isolation,Identification of Lactic Acid Degrading Bacteria in Alfalfa Silage and Their Degradation Characterization [J]. Biotechnology Bulletin, 2021, 37(9): 58-67. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||