








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (7): 26-36.doi: 10.13560/j.cnki.biotech.bull.1985.2022-1474
Previous Articles Next Articles
LI Yu1( ), LI Su-zhen2, CHEN Ru-mei2(
), LI Su-zhen2, CHEN Ru-mei2( ), LU Hai-qiang1(
), LU Hai-qiang1( )
)
Received:2022-12-02
Online:2023-07-26
Published:2023-08-17
Contact:
CHEN Ru-mei, LU Hai-qiang
E-mail:liyu397@126.com;chenrumei@caas.cn;luhaiqiang@hebau.edu.cn
LI Yu, LI Su-zhen, CHEN Ru-mei, LU Hai-qiang. Advances in the Regulation of Iron Homeostasis by bHLH Transcription Factors in Plant[J]. Biotechnology Bulletin, 2023, 39(7): 26-36.
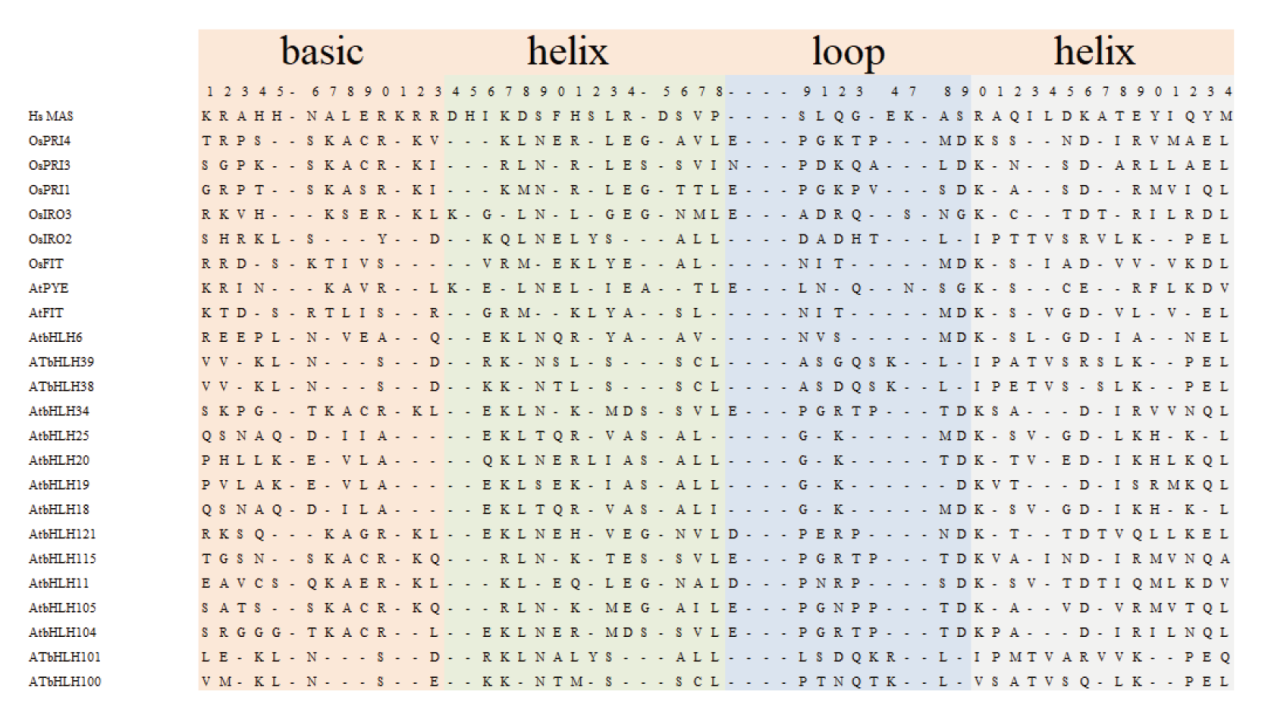
Fig. 1 Alignment of the bHLH domain of iron-regulated network main proteins The bHLH protein mentioned in the text and the typical human protein MAS with bHLH protein characteristics were selected in the figure. The shaded boxes represent the DNA binding alkaline region and two α spiral and a circular region
| [1] |
Wintz H, Fox T, Wu YY, et al. Expression profiles of Arabidopsis thaliana in mineral deficiencies reveal novel transporters involved in metal homeostasis[J]. J Biol Chem, 2003, 278(48): 47644-47653.
doi: 10.1074/jbc.M309338200 URL |
| [2] |
Briat JF, Dubos C, Gaymard F. Iron nutrition, biomass production, and plant product quality[J]. Trends Plant Sci, 2015, 20(1): 33-40.
doi: 10.1016/j.tplants.2014.07.005 URL |
| [3] |
Römheld V, Marschner H. Evidence for a specific uptake system for iron phytosiderophores in roots of grasses[J]. Plant Physiol, 1986, 80(1): 175-180.
doi: 10.1104/pp.80.1.175 pmid: 16664577 |
| [4] |
Kobayashi T, Nishizawa NK. Iron uptake, translocation, and regulation in higher plants[J]. Annu Rev Plant Biol, 2012, 63: 131-152.
doi: 10.1146/annurev-arplant-042811-105522 pmid: 22404471 |
| [5] |
Gao F, Dubos C. Transcriptional integration of plant responses to iron availability[J]. J Exp Bot, 2021, 72(6): 2056-2070.
doi: 10.1093/jxb/eraa556 pmid: 33246334 |
| [6] |
Fourcroy P, Tissot N, Gaymard F, et al. Facilitated Fe nutrition by phenolic compounds excreted by the Arabidopsis ABCG37/PDR9 transporter requires the IRT1/FRO2 high-affinity root Fe(2+)transport system[J]. Mol Plant, 2016, 9(3): 485-488.
doi: S1674-2052(15)00387-1 pmid: 26415695 |
| [7] |
Robinson NJ, Procter CM, Connolly EL, et al. A ferric-chelate reductase for iron uptake from soils[J]. Nature, 1999, 397(6721): 694-697.
doi: 10.1038/17800 URL |
| [8] |
Brumbarova T, Bauer P, Ivanov R. Molecular mechanisms governing Arabidopsis iron uptake[J]. Trends Plant Sci, 2015, 20(2): 124-133.
doi: 10.1016/j.tplants.2014.11.004 pmid: 25499025 |
| [9] |
Curie C, Panaviene Z, Loulergue C, et al. Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III)uptake[J]. Nature, 2001, 409(6818): 346-349.
doi: 10.1038/35053080 URL |
| [10] |
Zheng LQ, Ying YH, Wang L, et al. Identification of a novel iron regulated basic helix-loop-helix protein involved in Fe homeostasis in Oryza sativa[J]. BMC Plant Biol, 2010, 10: 166.
doi: 10.1186/1471-2229-10-166 |
| [11] |
Heim MA, Jakoby M, Werber M, et al. The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity[J]. Mol Biol Evol, 2003, 20(5): 735-747.
doi: 10.1093/molbev/msg088 pmid: 12679534 |
| [12] |
Li XX, Duan XP, Jiang HX, et al. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis[J]. Plant Physiol, 2006, 141(4): 1167-1184.
doi: 10.1104/pp.106.080580 URL |
| [13] |
Tarczewska A, Greb-Markiewicz B. The significance of the intrinsically disordered regions for the functions of the bHLH transcription factors[J]. Int J Mol Sci, 2019, 20(21): 5306.
doi: 10.3390/ijms20215306 URL |
| [14] |
Pires N, Dolan L. Origin and diversification of basic-helix-loop-helix proteins in plants[J]. Mol Biol Evol, 2010, 27(4): 862-874.
doi: 10.1093/molbev/msp288 pmid: 19942615 |
| [15] |
Hao YQ, Zong XM, Ren P, et al. Basic helix-loop-helix(bHLH)transcription factors regulate a wide range of functions in Arabidopsis[J]. Int J Mol Sci, 2021, 22(13): 7152.
doi: 10.3390/ijms22137152 URL |
| [16] |
Filiz E, Vatansever R, Ozyigit II. Dissecting a co-expression network of basic helix-loop-helix(bHLH)genes from phosphate(Pi)-starved soybean(Glycine max)[J]. Plant Gene, 2017, 9: 19-25.
doi: 10.1016/j.plgene.2016.12.001 URL |
| [17] |
Gao F, Robe K, Gaymard F, et al. The transcriptional control of iron homeostasis in plants: a tale of bHLH transcription factors?[J]. Front Plant Sci, 2019, 10: 6.
doi: 10.3389/fpls.2019.00006 pmid: 30713541 |
| [18] |
Gao F, Robe K, Bettembourg M, et al. The transcription factor bHLH121 interacts with bHLH105(ILR3)and its closest homologs to regulate iron homeostasis in Arabidopsis[J]. Plant Cell, 2020, 32(2): 508-524.
doi: 10.1105/tpc.19.00541 URL |
| [19] |
Selote D, Samira R, Matthiadis A, et al. Iron-binding E3 ligase mediates iron response in plants by targeting basic helix-loop-helix transcription factors[J]. Plant Physiol, 2015, 167(1): 273-286.
doi: 10.1104/pp.114.250837 pmid: 25452667 |
| [20] |
Rodríguez-Celma J, Connorton JM, Kruse I, et al. Arabidopsis BRUTUS-LIKE E3 ligases negatively regulate iron uptake by targeting transcription factor FIT for recycling[J]. Proc Natl Acad Sci USA, 2019, 116(35): 17584-17591.
doi: 10.1073/pnas.1907971116 pmid: 31413196 |
| [21] |
Wang N, Cui Y, Liu Y, et al. Requirement and functional redundancy of Ib subgroup bHLH proteins for iron deficiency responses and uptake in Arabidopsis thaliana[J]. Mol Plant, 2013, 6(2): 503-513.
doi: 10.1093/mp/sss089 URL |
| [22] | Akmakjian GZ, Riaz N, Guerinot ML. Photoprotection during iron deficiency is mediated by the bHLH transcription factors PYE and ILR3[J]. Proc Natl Acad Sci USA, 2021, 118(40): e2024918118. |
| [23] |
Liang G, Zhang HM, Li XL, et al. bHLH transcription factor bHLH115 regulates iron homeostasis in Arabidopsis thaliana[J]. J Exp Bot, 2017, 68(7): 1743-1755.
doi: 10.1093/jxb/erx043 pmid: 28369511 |
| [24] |
Li XL, Zhang HM, Ai Q, et al. Two bHLH transcription factors, bHLH34 and bHLH104, regulate iron homeostasis in Arabidopsis thaliana[J]. Plant Physiol, 2016, 170(4): 2478-2493.
doi: 10.1104/pp.15.01827 URL |
| [25] |
Lei RH, Li Y, Cai YR, et al. bHLH121 functions as a direct link that facilitates the activation of FIT by bHLH IVc transcription factors for maintaining Fe homeostasis in Arabidopsis[J]. Mol Plant, 2020, 13(4): 634-649.
doi: 10.1016/j.molp.2020.01.006 URL |
| [26] |
Gao F, Robe K, Dubos C. Further insights into the role of bHLH121 in the regulation of iron homeostasis in Arabidopsis thaliana[J]. Plant Signal Behav, 2020, 15(10): 1795582.
doi: 10.1080/15592324.2020.1795582 URL |
| [27] |
Schwarz B, Bauer P. FIT, a regulatory hub for iron deficiency and stress signaling in roots, and FIT-dependent and-independent gene signatures[J]. J Exp Bot, 2020, 71(5): 1694-1705.
doi: 10.1093/jxb/eraa012 pmid: 31922570 |
| [28] |
Bauer P, Ling HQ, Guerinot ML. Fit, the fer-like iron deficiency induced transcription factor in Arabidopsis[J]. Plant Physiol Biochem, 2007, 45(5): 260-261.
doi: 10.1016/j.plaphy.2007.03.006 URL |
| [29] | Sivitz AB, Hermand V, Curie C, et al. Arabidopsis bHLH100 and bHLH101 control iron homeostasis via a FIT-independent pathway[J]. PLoS One, 2012, 7(9): e44843. |
| [30] | Yuan YX, Wu HL, Wang N, et al. FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis[J]. Cell Res, 2008, 18(3): 385-397. |
| [31] |
Long TA, Tsukagoshi H, Busch W, et al. The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots[J]. Plant Cell, 2010, 22(7): 2219-2236.
doi: 10.1105/tpc.110.074096 URL |
| [32] |
Tanabe N, Noshi M, Mori D, et al. The basic helix-loop-helix transcription factor, bHLH11 functions in the iron-uptake system in Arabidopsis thaliana[J]. J Plant Res, 2019, 132(1): 93-105.
doi: 10.1007/s10265-018-1068-z |
| [33] |
Gratz R, Manishankar P, Ivanov R, et al. CIPK11-dependent phosphorylation modulates FIT activity to promote Arabidopsis iron acquisition in response to calcium signaling[J]. Dev Cell, 2019, 48(5): 726-740.e10.
doi: 10.1016/j.devcel.2019.01.006 URL |
| [34] |
Gratz R, Brumbarova T, Ivanov R, et al. Phospho-mutant activity assays provide evidence for alternative phospho-regulation pathways of the transcription factor fer-like iron deficiency-induced transcription factor[J]. New Phytol, 2020, 225(1): 250-267.
doi: 10.1111/nph.16168 pmid: 31487399 |
| [35] |
Cui Y, Chen CL, Cui M, et al. Four IVa bHLH transcription factors are novel interactors of FIT and mediate JA inhibition of iron uptake in Arabidopsis[J]. Mol Plant, 2018, 11(9): 1166-1183.
doi: 10.1016/j.molp.2018.06.005 URL |
| [36] |
Matsuoka K, Furukawa J, Bidadi H, et al. Gibberellin-induced expression of Fe uptake-related genes in Arabidopsis[J]. Plant Cell Physiol, 2014, 55(1): 87-98.
doi: 10.1093/pcp/pct160 pmid: 24192296 |
| [37] |
Wild M, Davière JM, Regnault T, et al. Tissue-specific regulation of gibberellin signaling fine-tunes Arabidopsis iron-deficiency responses[J]. Dev Cell, 2016, 37(2): 190-200.
doi: 10.1016/j.devcel.2016.03.022 URL |
| [38] |
Chao Q, Rothenberg M, Solano R, et al. Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins[J]. Cell, 1997, 89(7): 1133-1144.
doi: 10.1016/s0092-8674(00)80300-1 pmid: 9215635 |
| [39] |
Lingam S, Mohrbacher J, Brumbarova T, et al. Interaction between the bHLH transcription factor FIT and ETHYLENE INSENSITIVE3/ETHYLENE INSENSITIVE3-LIKE1 reveals molecular linkage between the regulation of iron acquisition and ethylene signaling in Arabidopsis[J]. Plant Cell, 2011, 23(5): 1815-1829.
doi: 10.1105/tpc.111.084715 URL |
| [40] |
Yang Y, Ou B, Zhang JZ, et al. The Arabidopsis mediator subunit MED16 regulates iron homeostasis by associating with EIN3/EIL1 through subunit MED25[J]. Plant J, 2014, 77(6): 838-851.
doi: 10.1111/tpj.2014.77.issue-6 URL |
| [41] |
Zhang Y, Wu HL, Wang N, et al. Mediator subunit 16 functions in the regulation of iron uptake gene expression in Arabidopsis[J]. New Phytol, 2014, 203(3): 770-783.
doi: 10.1111/nph.12860 pmid: 24889527 |
| [42] |
Ishimaru Y, Suzuki M, Tsukamoto T, et al. Rice plants take up iron as an Fe3+-phytosiderophore and as Fe2+[J]. Plant J, 2006, 45(3): 335-346.
doi: 10.1111/j.1365-313X.2005.02624.x pmid: 16412081 |
| [43] |
Zhang HM, Li Y, Pu MN, et al. Oryza sativa positive regulator of iron deficiency response 2(ospri2)and ospri3 are involved in the maintenance of fe homeostasis[J]. Plant Cell Environ, 2020, 43(1): 261-274.
doi: 10.1111/pce.v43.1 URL |
| [44] |
Kobayashi T, Nagasaka S, Senoura T, et al. Iron-binding haemerythrin RING ubiquitin ligases regulate plant iron responses and accumulation[J]. Nat Commun, 2013, 4: 2792.
doi: 10.1038/ncomms3792 pmid: 24253678 |
| [45] |
Zhang HM, Li Y, Yao XN, et al. POSITIVE REGULATOR OF IRON HOMEOSTASIS1, OsPRI1, facilitates iron homeostasis[J]. Plant Physiol, 2017, 175(1): 543-554.
doi: 10.1104/pp.17.00794 pmid: 28751317 |
| [46] | Wang WJ, Ye J, Ma YR, et al. OsIRO3 plays an essential role in iron deficiency responses and regulates iron homeostasis in rice[J]. Plants(Basel), 2020, 9(9): 1095. |
| [47] |
Wang WJ, Ye J, Xu H, et al. osbhlh061 links topless/topless-related repressor proteins with positive regulator of iron homeostasis 1 to maintain iron homeostasis in rice[J]. New Phytol, 2022, 234(5): 1753-1769.
doi: 10.1111/nph.18096 pmid: 35288933 |
| [48] |
Ogo Y, Itai RN, Kobayashi T, et al. OsIRO2 is responsible for iron utilization in rice and improves growth and yield in calcareous soil[J]. Plant Mol Biol, 2011, 75(6): 593-605.
doi: 10.1007/s11103-011-9752-6 pmid: 21331630 |
| [49] |
Ogo Y, Itai RN, Nakanishi H, et al. The rice bHLH protein OsIRO2 is an essential regulator of the genes involved in Fe uptake under Fe-deficient conditions[J]. Plant J, 2007, 51(3): 366-377.
doi: 10.1111/j.1365-313X.2007.03149.x pmid: 17559517 |
| [50] |
Li CY, Li Y, Xu P, et al. OsIRO3 negatively regulates Fe homeostasis by repressing the expression of OsIRO2[J]. Plant J, 2022, 111(4): 966-978.
doi: 10.1111/tpj.v111.4 URL |
| [51] |
Wang SD, Li L, Ying YH, et al. A transcription factor OsbHLH156 regulates Strategy II iron acquisition through localising IRO2 to the nucleus in rice[J]. New Phytol, 2020, 225(3): 1247-1260.
doi: 10.1111/nph.16232 pmid: 31574173 |
| [52] | Trofimov K, Ivanov R, Eutebach M, et al. Mobility and localization of the iron deficiency-induced transcription factor bHLH039 change in the presence of FIT[J]. Plant Direct, 2019, 3(12): e00190. |
| [53] |
Brumbarova T, Bauer P. Iron-mediated control of the basic helix-loop-helix protein FER, a regulator of iron uptake in tomato[J]. Plant Physiol, 2005, 137(3): 1018-1026.
doi: 10.1104/pp.104.054270 pmid: 15695640 |
| [54] |
Bereczky Z, Wang HY, Schubert V, et al. Differential regulation of nramp and irt metal transporter genes in wild type and iron uptake mutants of tomato[J]. J Biol Chem, 2003, 278(27): 24697-24704.
doi: 10.1074/jbc.M301365200 pmid: 12709425 |
| [55] |
Du J, Huang ZA, Wang B, et al. SlbHLH068 interacts with FER to regulate the iron-deficiency response in tomato[J]. Ann Bot, 2015, 116(1): 23-34.
doi: 10.1093/aob/mcv058 URL |
| [56] |
Zhao Q, Ren YR, Wang QJ, et al. Overexpression of MdbHLH104 gene enhances the tolerance to iron deficiency in apple[J]. Plant Biotechnol J, 2016, 14(7): 1633-1645.
doi: 10.1111/pbi.2016.14.issue-7 URL |
| [57] |
Xu HM, Wang Y, Chen F, et al. Isolation and characterization of the iron-regulated MxbHLH01 gene in Malus xiaojinensis[J]. Plant Mol Biol Rep, 2011, 29(4): 936-942.
doi: 10.1007/s11105-011-0305-6 URL |
| [58] |
Yin L, Wang Y, Yan M, et al. Molecular cloning, polyclonal antibody preparation, and characterization of a functional iron-related transcription factor IRO2 from Malus xiaojinensis[J]. Plant Physiol Biochem, 2013, 67: 63-70.
doi: 10.1016/j.plaphy.2013.02.021 URL |
| [59] | Tian Y, Pu XD, Yu HY, et al. Genome-wide characterization and analysis of bHLH transcription factors related to crocin biosynthesis in Gardenia jasminoides Ellis(Rubiaceae)[J]. Biomed Res Int, 2020, 2020: 2903861. |
| [60] |
Ling HQ, Bauer P, Bereczky Z, et al. The tomato fer gene encoding a bHLH protein controls iron-uptake responses in roots[J]. Proc Natl Acad Sci USA, 2002, 99(21): 13938-13943.
doi: 10.1073/pnas.212448699 URL |
| [1] | WANG Zi-ying, LONG Chen-jie, FAN Zhao-yu, ZHANG Lei. Screening of OsCRK5-interacted Proteins in Rice Using Yeast Two-hybrid System [J]. Biotechnology Bulletin, 2023, 39(9): 117-125. |
| [2] | WU Yuan-ming, LIN Jia-yi, LIU Yu-xi, LI Dan-ting, ZHANG Zong-qiong, ZHENG Xiao-ming, PANG Hong-bo. Identification of Rice Plant Height-associated QTL Using BSA-seq and RNA-seq [J]. Biotechnology Bulletin, 2023, 39(8): 173-184. |
| [3] | YAO Sha-sha, WANG Jing-jing, WANG Jun-jie, LIANG Wei-hong. Molecular Mechanisms of Rice Grain Size Regulation Related to Plant Hormone Signaling Pathways [J]. Biotechnology Bulletin, 2023, 39(8): 80-90. |
| [4] | LI Zhi-qi, YUAN Yue, MIAO Rong-qing, PANG Qiu-ying, ZHANG Ai-qin. Melatonin Contents in Eutrema salsugineum and Arabidopsis thaliana Under Salt Stress, and Expression Pattern Analysis of Synthesis Related Genes [J]. Biotechnology Bulletin, 2023, 39(5): 142-151. |
| [5] | LIANG Cheng-gang, WANG Yan, LI Tian, OHSUGI Ryu, AOKI Naohiro. Effect of SP1 on Panicle Architecture by Regulating Carbohydrate Remobilization [J]. Biotechnology Bulletin, 2023, 39(5): 152-159. |
| [6] | ZHOU Ding-ding, LI Hui-hu, TANG Xing-yong, YU Fa-xin, KONG Dan-yu, LIU Yi. Research Progress in the Biosynthesis and Regulation of Glycyrrhizic Acid and Liquiritin [J]. Biotechnology Bulletin, 2023, 39(5): 44-53. |
| [7] | YANG Mao, LIN Yu-feng, DAI Yang-shuo, PAN Su-jun, PENG Wei-ye, YAN Ming-xiong, LI Wei, WANG Bing, DAI Liang-ying. OsDIS1 Negatively Regulates Rice Drought Tolerance Through Antioxidant Pathways [J]. Biotechnology Bulletin, 2023, 39(2): 88-95. |
| [8] | JIANG Min-xuan, LI Kang, LUO Liang, LIU Jian-xiang, LU Hai-ping. Advances on the Expressions of Foreign Proteins in Plants [J]. Biotechnology Bulletin, 2023, 39(11): 110-122. |
| [9] | LIN Rong, ZHENG Yue-ping, XU Xue-zhen, LI Dan-dan, ZHENG Zhi-fu. Functional Analysis of ACOL8 Gene in the Ethylene Synthesis and Response in Arabidopsis thaliana [J]. Biotechnology Bulletin, 2023, 39(1): 157-165. |
| [10] | JIANG Nan, SHI Yang, ZHAO Zhi-hui, LI Bin, ZHAO Yi-hui, YANG Jun-biao, YAN Jia-ming, JIN Yu-fan, CHEN Ji, HUANG Jin. Expression and Functional Analysis of OsPT1 Gene in Rice Under Cadmium Stress [J]. Biotechnology Bulletin, 2023, 39(1): 166-174. |
| [11] | CHEN Guang, LI Jia, DU Rui-ying, WANG Xu. Identification and Gene Functional Analysis of Salinity-hypersensitive Mutant ss2 in Rice [J]. Biotechnology Bulletin, 2022, 38(9): 158-166. |
| [12] | GAO Xiao-rong, DING Yao, LV Jun. Effects of Pseudomonas sp. PR3,a Pyrene-degrading Bacterium with Plant Growth-promoting Properties,on Rice Growth Under Pyrene Stress [J]. Biotechnology Bulletin, 2022, 38(9): 226-236. |
| [13] | HUANG Jing, ZHU Liang, XUE Peng-bo, FU Qiang. Research on Mechanism and QTL Mapping Associated with Cadmium Accumulation in Rice Leaves and Grains [J]. Biotechnology Bulletin, 2022, 38(8): 118-126. |
| [14] | CHEN Guang, LI Jia, DU Rui-ying, WANG Xu. pOsHAK1:OsFLN2 Expression Enhances the Drought Tolerance by Altering Sugar Metabolism in Rice [J]. Biotechnology Bulletin, 2022, 38(8): 92-100. |
| [15] | LI Bai, CAI Zhi-jun, WANG Lei, CHEN Jie, CAO Kui-rong, LI Jun, CHONG Gao-jun. Development and Application of the Combinatorial Marker for the Rice Blast Resistance Gene Pigm [J]. Biotechnology Bulletin, 2022, 38(7): 153-159. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||