








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (8): 213-219.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0128
Previous Articles Next Articles
XU Jing1( ), ZHU Hong-lin1, LIN Yan-hui1, TANG Li-qiong1, TANG Qing-jie1,2, WANG Xiao-ning1,2(
), ZHU Hong-lin1, LIN Yan-hui1, TANG Li-qiong1, TANG Qing-jie1,2, WANG Xiao-ning1,2( )
)
Received:2023-02-15
Online:2023-08-26
Published:2023-09-05
Contact:
WANG Xiao-ning
E-mail:xujing6732807@126.com;wxning2599@163.com
XU Jing, ZHU Hong-lin, LIN Yan-hui, TANG Li-qiong, TANG Qing-jie, WANG Xiao-ning. Cloning of IbHQT1 Promoter and Identification of Upstream Regulatory Factors in Sweet Potato[J]. Biotechnology Bulletin, 2023, 39(8): 213-219.
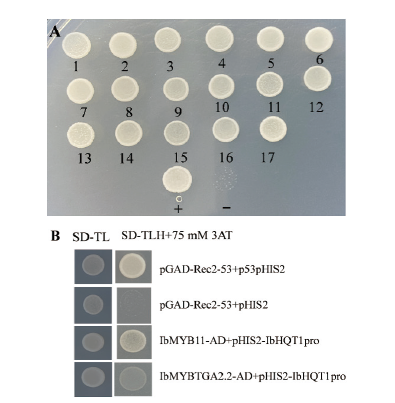
Fig. 4 Screening and identification of IbHQT1 promoter interacting proteins A: Secondary screening of binding protein interacting with IbHQT1 promoter.1-17: Yeast colonies; +: positive control; -:negative control. B: The interaction of IbHQT1 promoter with IbMYB11 and IbTGA2.2
| [1] |
Tajik N, Tajik M, Mack I, et al. The potential effects of chlorogenic acid, the main phenolic components in coffee, on health: a comprehensive review of the literature[J]. Eur J Nutr, 2017, 56(7): 2215-2244.
doi: 10.1007/s00394-017-1379-1 pmid: 28391515 |
| [2] |
Naveed M, Hejazi V, Abbas M, et al. Chlorogenic acid(CGA): A pharmacological review and call for further research[J]. Biomed Pharmacother, 2018, 97: 67-74.
doi: 10.1016/j.biopha.2017.10.064 URL |
| [3] |
Wianowska D, Gil M. Recent advances in extraction and analysis procedures of natural chlorogenic acids[J]. Phytochem Rev, 2019, 18(1): 273-302.
doi: 10.1007/s11101-018-9592-y |
| [4] |
Mohanraj R, Sivasankar S. Sweet potato(Ipomoea batatas[L.]Lam): a valuable medicinal food: a review[J]. J Med Food, 2014, 17(7): 733-741.
doi: 10.1089/jmf.2013.2818 pmid: 24921903 |
| [5] |
Tanaka M, Ishiguro K, Oki T, et al. Functional components in sweetpotato and their genetic improvement[J]. Breed Sci, 2017, 67(1): 52-61.
doi: 10.1270/jsbbs.16125 URL |
| [6] | Islam MS, Yoshimoto M, Yamakawa O. Distribution and physiological functions of caffeoylquinic acid derivatives in leaves of sweetpotato genotypes[J]. J Food Sci, 2006, 68(1): 111-116. |
| [7] |
Rautenbach F, Faber M, Laurie S, et al. Antioxidant capacity and antioxidant content in roots of 4 sweetpotato varieties[J]. J Food Sci, 2010, 75(5): C400-C405.
doi: 10.1111/jfds.2010.75.issue-5 URL |
| [8] |
李佳银, 于欢, 石伯阳, 等. 甘薯茎叶中异槲皮苷及咖啡酰基奎宁酸类衍生物的抗氧化活性[J]. 食品科学, 2013, 34(7): 111-114.
doi: 10.7506/spkx1002-6630-201307023 |
| Li JY, Yu H, Shi BY, et al. Antioxidant activity of isoquercitrin and caffeoylquinic acid derivatives from sweet potato stems and leaves[J]. Food sci, 2013, 34(7): 111-114. | |
| [9] | 刘雪辉, 李觅路, 谭斌, 等. 紫甘薯茎叶中绿原酸及异绿原酸对α-葡萄糖苷酶的抑制作用[J]. 现代食品科技, 2014, 30(3): 103-107. |
| Liu XH, Li ML, Tan B, et al. Inhibitory effects of chlorogenic acid and isochlorogenic acid from purple sweet potato leaves on α-glucosidase[J]. Mod Food Sci Technol, 2014, 30(3): 103-107. | |
| [10] | 王伟, 阮妙鸿, 邱永祥, 等. 甘薯抗薯瘟病的苯丙烷类代谢研究[J]. 中国生态农业学报, 2009, 17(5): 944-948. |
|
Wang W, Ruan MH, Qiu YX, et al. Phenylaprapanoid metabolism of sweet potato against Pseudomonas solanacearum[J]. Chin J Eco Agric, 2009, 17(5): 944-948.
doi: 10.3724/SP.J.1011.2009.00944 URL |
|
| [11] | 刘美艳, 孙厚俊, 王景景, 等. 甘薯块根抗黑斑病酚类物质代谢的研究[J]. 中国农学通报, 2012, 28(24): 226-230. |
| Liu MY, Sun HJ, Wang JJ, et al. The study on phenol metabolism in sweet potato infected by Ceratocystis fimbriata Ellis et Halsted[J]. Chin Agric Sci Bull, 2012, 28(24): 226-230. | |
| [12] |
刘海晶, 段立清, 李海平, 等. 绿原酸提高舞毒蛾核型多角体病毒(LdNPV)的致病力[J]. 昆虫学报, 2016, 59(5): 568-572.
doi: 10.16380/j.kcxb.2016.05.012 |
| Liu HJ, Duan LQ, Li HP, et al. Chlorogenic acid enhances the virulence of Lymantria dispar necleopolyhydrovirus(LdNPV)[J]. Acta Entomol Sin, 2016, 59(5): 568-572. | |
| [13] | 王珍, 李宗芸. 绿原酸的生物活性及甘薯绿原酸研究进展[J]. 江苏师范大学学报: 自然科学版, 2017, 35(3): 30-34, 48. |
| Wang Z, Li ZY. Research progress of biological activity of chlorogenic acid and chlorogenic acid in sweetpotato[J]. J Jiangsu Norm Univ Nat Sci Ed, 2017, 35(3): 30-34, 48. | |
| [14] |
Liu Y, Su WJ, Wang LJ, et al. Integrated transcriptome, small RNA and degradome sequencing approaches proffer insights into chlorogenic acid biosynthesis in leafy sweet potato[J]. PLoS One, 2021, 16(1): e0245266.
doi: 10.1371/journal.pone.0245266 URL |
| [15] |
Liu Q, Liu Y, Xu YC, et al. Overexpression of and RNA interference with hydroxycinnamoyl-CoA quinate hydroxycinnamoyl transferase affect the chlorogenic acid metabolic pathway and enhance salt tolerance in Taraxacum antungense Kitag[J]. Phytochem Lett, 2018, 28: 116-123.
doi: 10.1016/j.phytol.2018.10.003 URL |
| [16] |
Moglia A, Lanteri S, Comino C, et al. Dual catalytic activity of hydroxycinnamoyl-coenzyme A quinate transferase from tomato allows it to moonlight in the synthesis of both mono- and dicaffeoylquinic acids[J]. Plant Physiol, 2014, 166(4): 1777-1787.
doi: 10.1104/pp.114.251371 pmid: 25301886 |
| [17] |
Payyavula RS, Shakya R, Sengoda VG, et al. Synthesis and regulation of chlorogenic acid in potato: Rerouting phenylpropanoid flux in HQT-silenced lines[J]. Plant Biotechnol J, 2015, 13(4): 551-564.
doi: 10.1111/pbi.12280 pmid: 25421386 |
| [18] |
Zhang JR, Wu ML, Li WD, et al. Regulation of chlorogenic acid biosynthesis by hydroxycinnamoyl CoA quinate hydroxycinnamoyl transferase in Lonicera japonica[J]. Plant Physiol Biochem, 2017, 121: 74-79.
doi: 10.1016/j.plaphy.2017.10.017 URL |
| [19] | Moglia A, Acquadro A, Eljounaidi K, et al. Genome-wide identification of BAHD acyltransferases and in vivo characterization of HQT-like enzymes involved in caffeoylquinic acid synthesis in globe artichoke[J]. Front Plant Sci, 2016, 7: 1424. |
| [20] |
Mudau SP, Steenkamp PA, Piater LA, et al. Metabolomics-guided investigations of unintended effects of the expression of the hydroxycinnamoyl quinate hydroxycinnamoyltransferase(hqt1)gene from Cynara cardunculus var. scolymus in Nicotiana tabacum cell cultures[J]. Plant Physiol Biochem, 2018, 127: 287-298.
doi: 10.1016/j.plaphy.2018.04.005 URL |
| [21] |
Li YQ, Kong DX, Bai M, et al. Correlation of the temporal and spatial expression patterns of HQT with the biosynthesis and accumulation of chlorogenic acid in Lonicera japonica flowers[J]. Hortic Res, 2019, 6: 73.
doi: 10.1038/s41438-019-0154-2 |
| [22] |
Patra B, Schluttenhofer C, Wu YM, et al. Transcriptional regulation of secondary metabolite biosynthesis in plants[J]. Biochim Biophys Acta, 2013, 1829(11): 1236-1247.
doi: 10.1016/j.bbagrm.2013.09.006 pmid: 24113224 |
| [23] |
Chezem WR, Clay NK. Regulation of plant secondary metabolism and associated specialized cell development by MYBs and bHLHs[J]. Phytochemistry, 2016, 131: 26-43.
doi: S0031-9422(16)30158-3 pmid: 27569707 |
| [24] | 赵莹, 杨欣宇, 赵晓丹, 等. 植物类黄酮化合物生物合成调控研究进展[J]. 食品工业科技, 2021, 42(21): 454-463. |
| Zhao Y, Yang XY, Zhao XD, et al. Research progress on regulation of plant flavonoids biosynthesis[J]. Sci Technol Food Ind, 2021, 42(21): 454-463. | |
| [25] |
Xu J, Zhu JH, Lin YH, et al. Comparative transcriptome and weighted correlation network analyses reveal candidate genes involved in chlorogenic acid biosynthesis in sweet potato[J]. Sci Rep, 2022, 12(1): 1-11.
doi: 10.1038/s41598-021-99269-x |
| [26] |
Zhou ML, Memelink J. Jasmonate-responsive transcription factors regulating plant secondary metabolism[J]. Biotechnol Adv, 2016, 34(4): 441-449.
doi: S0734-9750(16)30011-8 pmid: 26876016 |
| [27] | 段乐鹏, 谢丽琼, 马艺沔, 等. 转录因子对药用植物次生代谢的调控作用[J]. 中药材, 2021, 44(4): 1002-1007. |
| Duan LP, Xie LQ, Ma YG, et al. Regulation of transcription factors on secondary metabolism of medicinal plants[J]. J Chin Med Mater, 2021, 44(4): 1002-1007. | |
| [28] |
Ma DW, Constabel CP. MYB repressors as regulators of phenylpropanoid metabolism in plants[J]. Trends Plant Sci, 2019, 24(3): 275-289
doi: S1360-1385(18)30290-5 pmid: 30704824 |
| [29] |
Anwar M, Chen L, Xiao YB, et al. Recent advanced metabolic and genetic engineering of phenylpropanoid biosynthetic pathways[J]. Int J Mol Sci, 2021, 22(17): 9544.
doi: 10.3390/ijms22179544 URL |
| [30] |
Duan AQ, Tan SS, Deng YJ, et al. Genome-wide identification and evolution analysis of R2R3-MYB gene family reveals S6 subfamily R2R3-MYB transcription factors involved in anthocyanin biosynthesis in carrot[J]. Int J Mol Sci, 2022, 23: 11859.
doi: 10.3390/ijms231911859 URL |
| [31] |
Zhao L, Song ZB, Wang BW, et al. R2R3-MYB transcription factor NtMYB330 regulates proanthocyanidin biosynthesis and seed germination in tobacco(Nicotiana tabacum L.)[J]. Front Plant Sci, 2022, 12: 819247.
doi: 10.3389/fpls.2021.819247 URL |
| [32] |
Tian Q, Han LM, Zhu XY, et al. SmMYB4 is a R2R3-MYB transcriptional repressor regulating the biosynthesis of phenolic acids and tanshinones in Salvia miltiorrhiza[J]. Metabolites, 2022, 12(10): 968.
doi: 10.3390/metabo12100968 URL |
| [33] |
Stracke R, Jahns O, Keck M, et al. Analysis of production of flavonol glycosides-dependent flavonol glycoside accumulation in Arabidopsis thaliana plants reveals myb11-, myb12- and myb111-independent flavonol glycoside accumulation[J]. New Phytol, 2010, 188(4): 985-1000.
doi: 10.1111/j.1469-8137.2010.03421.x pmid: 20731781 |
| [34] | Li Y, Chen M, Wang SL, et al. AtMYB11 regulates caffeoylquinic acid and flavonol synthesis in tomato and tobacco[J]. Plant Cell Tissue Organ Cult PCTOC, 2015, 122(2): 309-319. |
| [35] |
Luo J, Butelli E, Hill L, et al. AtMYB12 regulates caffeoyl quinic acid and flavonol synthesis in tomato: expression in fruit results in very high levels of both types of polyphenol[J]. Plant J, 2008, 56(2): 316-326.
doi: 10.1111/j.1365-313X.2008.03597.x URL |
| [36] |
Guo D, Li HL, Zhu JH, et al. HbTGA1, a TGA transcription factor from Hevea brasiliensis, regulates the expression of multiple natural rubber biosynthesis genes[J]. Front Plant Sci, 2022, 13: 909098.
doi: 10.3389/fpls.2022.909098 URL |
| [37] |
Huo YB, Zhang B, Chen L, et al. Isolation and functional characterization of the promoters of miltiradiene synthase genes, TwTPS27a and TwTPS27b, and interaction analysis with the transcription factor TwTGA1 from Tripterygium wilfordii[J]. Plants, 2021, 10(2): 418.
doi: 10.3390/plants10020418 URL |
| [38] |
Lyu ZY, Guo ZY, Zhang LD, et al. Interaction of BZIP transcription factor TGA6 with salicylic acid signaling modulates artemisinin biosynthesis in Artemisia annua[J]. J Exp Bot, 2019, 70(15): 3969-3979.
doi: 10.1093/jxb/erz166 URL |
| [39] |
Han J, Liu HT, Wang SC, et al. A class I TGA transcription factor from Tripterygium wilfordii Hook. f. modulates the biosynthesis of secondary metabolites in both native and heterologous hosts[J]. Plant Sci, 2020, 290: 110293.
doi: 10.1016/j.plantsci.2019.110293 URL |
| [40] |
Mao ZL, Jiang HY, Wang S, et al. The MdHY5-MdWRKY41-MdMYB transcription factor cascade regulates the anthocyanin and proanthocyanidin biosynthesis in red-fleshed apple[J]. Plant Sci, 2021, 306: 110848.
doi: 10.1016/j.plantsci.2021.110848 URL |
| [41] |
Sun QG, Jiang SH, Zhang TL, et al. Apple NAC transcription factor MdNAC52 regulates biosynthesis of anthocyanin and proanthocyanidin through MdMYB9 and MdMYB11[J]. Plant Sci, 2019, 289: 110286.
doi: 10.1016/j.plantsci.2019.110286 URL |
| [42] |
An JP, Yao JF, Xu RR, et al. Apple bZIP transcription factor MdbZIP44 regulates abscisic acid-promoted anthocyanin accumulation[J]. Plant Cell Environ, 2018, 41(11): 2678-2692.
doi: 10.1111/pce.v41.11 URL |
| [43] |
Xu WJ, Dubos C, Lepiniec L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes[J]. Trends Plant Sci, 2015, 20(3): 176-185.
doi: 10.1016/j.tplants.2014.12.001 pmid: 25577424 |
| [1] | HUANG Xiao-long, SUN Gui-lian, MA Dan-dan, YAN Hui-qing. Construction of Yeast One-hybrid Library and Screening of Factors Regulating LAZY1 Expression in Rice [J]. Biotechnology Bulletin, 2023, 39(9): 126-135. |
| [2] | HAN Hao-zhang, ZHANG Li-hua, LI Su-hua, ZHAO Rong, WANG Fang, WANG Xiao-li. Construction of cDNA Library of Cinnamomun bodinieri Induced by Saline-alkali Stress and Screening of CbP5CS Upstream Regulators [J]. Biotechnology Bulletin, 2023, 39(9): 236-245. |
| [3] | LYU Qiu-yu, SUN Pei-yuan, RAN Bin, WANG Jia-rui, CHEN Qing-fu, LI Hong-you. Cloning, Subcellular Localization and Expression Analysis of the Transcription Factor Gene FtbHLH3 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 194-203. |
| [4] | LI Bo, LIU He-xia, CHEN Yu-ling, ZHOU Xing-wen, ZHU Yu-lin. Cloning, Subcellular Localization and Expression Analysis of CnbHLH79 Transcription Factor from Camellia nitidissima [J]. Biotechnology Bulletin, 2023, 39(8): 241-250. |
| [5] | CHEN Xiao, YU Ming-lan, WU Long-kun, ZHENG Xiao-ming, PANG Hong-bo. Research Progress in lncRNA and Their Responses to Low Temperature Stress in Plant [J]. Biotechnology Bulletin, 2023, 39(7): 1-12. |
| [6] | GUO Yi-ting, ZHAO Wen-ju, REN Yan-jing, ZHAO Meng-liang. Identification and Analysis of NAC Transcription Factor Family Genes in Helianthus tuberosus L. [J]. Biotechnology Bulletin, 2023, 39(6): 217-232. |
| [7] | FENG Shan-shan, WANG Lu, ZHOU Yi, WANG You-ping, FANG Yu-jie. Research Progresses on WOX Family Genes in Regulating Plant Development and Abiotic Stress Response [J]. Biotechnology Bulletin, 2023, 39(5): 1-13. |
| [8] | WANG Bing, ZHAO Hui-na, YU Jing, YU Shi-zhou, LEI Bo. Research Progress in the Regulation of Plant Branch Development [J]. Biotechnology Bulletin, 2023, 39(5): 14-22. |
| [9] | ZHANG Xin-bo, CUI Hao-liang, SHI Pei-hua, GAO Jin-chun, ZHAO Shun-ran, TAO Chen-yu. Research Progress in Low-input Chromatin Immunoprecipitation Assay [J]. Biotechnology Bulletin, 2023, 39(4): 227-235. |
| [10] | GE Yan-rui, ZHAO Ran, XU Jing, LI Ruo-fan, HU Yun-tao, LI Rui-li. Advances in the Development and Regulation of Vascular Cambium [J]. Biotechnology Bulletin, 2023, 39(3): 13-25. |
| [11] | LIU Cheng-xia, SUN Zong-yan, LUO Yun-bo, ZHU Hong-liang, QU Gui-qin. Multifaceted Roles of bHLH Phosphorylation in Regulation of Plant Physiological Functions [J]. Biotechnology Bulletin, 2023, 39(3): 26-34. |
| [12] | ZHAO Meng-liang, GUO Yi-ting, REN Yan-jing. Identification and Analysis of WRKY Transcription Factor Family Genes in Helianthus tuberosus [J]. Biotechnology Bulletin, 2023, 39(2): 116-125. |
| [13] | HAN Fang-ying, HU Xin, WANG Nan-nan, XIE Yu-hong, WANG Xiao-yan, ZHU Qiang. Research Progress in Response of DREBs to Abiotic Stress in Plant [J]. Biotechnology Bulletin, 2023, 39(11): 86-98. |
| [14] | CHEN Chu-yi, YANG Xiao-mei, CHEN Sheng-yan, CHEN Bin, YUE Li-ran. Expression Analysis of the ZF-HD Gene Family in Chrysanthemum nankingense Under Drought and ABA Treatment [J]. Biotechnology Bulletin, 2023, 39(11): 270-282. |
| [15] | FENG Ce-ting, JIANG Lyu, LIU Xing-ying, LUO Le, PAN Hui-tang, ZHANG Qi-xiang, YU Chao. Identification of the NAC Gene Family in Rosa persica and Response Analysis Under Drought Stress [J]. Biotechnology Bulletin, 2023, 39(11): 283-296. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||