








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (7): 323-334.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0076
Previous Articles Next Articles
TIAN Tong-tong1,2( ), GE Jia-zhen1, GAO Peng-cheng1, LI Xue-rui1, SONG Guo-dong1, ZHENG Fu-ying1(
), GE Jia-zhen1, GAO Peng-cheng1, LI Xue-rui1, SONG Guo-dong1, ZHENG Fu-ying1( ), CHU Yue-feng1,2(
), CHU Yue-feng1,2( )
)
Received:2024-01-17
Online:2024-07-26
Published:2024-07-30
Contact:
ZHENG Fu-ying, CHU Yue-feng
E-mail:t1123859519@163.com;zhengfuying@caas.cn;chuyuefeng@caas.cn
TIAN Tong-tong, GE Jia-zhen, GAO Peng-cheng, LI Xue-rui, SONG Guo-dong, ZHENG Fu-ying, CHU Yue-feng. Whole Genome Sequencing and Bioinformatics Analysis of Mycoplasma ovipneumoniae GH3-3 Strain[J]. Biotechnology Bulletin, 2024, 40(7): 323-334.
| 菌株 Strain | 基因组大小 Genome size/bp | 编码基因数量 Gene number/个 | GC含量 GC content/% |
|---|---|---|---|
| GH 3-3株 | 1 060 772 | 730 | 29.66 |
| NXNK2203株 | 1 014 835 | 686 | 29.23 |
| NM2010株 | 1 084 159 | 888 | 29.14 |
| HHHT2017株 | 1 019 689 | 807 | 29.068 |
| SC01株 | 1 020 601 | 864 | 28.85 |
| WXB1株 | 1 084 159 | 888 | 29.14 |
| 150株 | 1 053 380 | 714 | 29.15 |
| 274株 | 1 081 404 | 747 | 28.82 |
| Y98株 | 1 021 486 | 819 | 29.21 |
Table 1 Comparison of genomic information of different MO strains
| 菌株 Strain | 基因组大小 Genome size/bp | 编码基因数量 Gene number/个 | GC含量 GC content/% |
|---|---|---|---|
| GH 3-3株 | 1 060 772 | 730 | 29.66 |
| NXNK2203株 | 1 014 835 | 686 | 29.23 |
| NM2010株 | 1 084 159 | 888 | 29.14 |
| HHHT2017株 | 1 019 689 | 807 | 29.068 |
| SC01株 | 1 020 601 | 864 | 28.85 |
| WXB1株 | 1 084 159 | 888 | 29.14 |
| 150株 | 1 053 380 | 714 | 29.15 |
| 274株 | 1 081 404 | 747 | 28.82 |
| Y98株 | 1 021 486 | 819 | 29.21 |
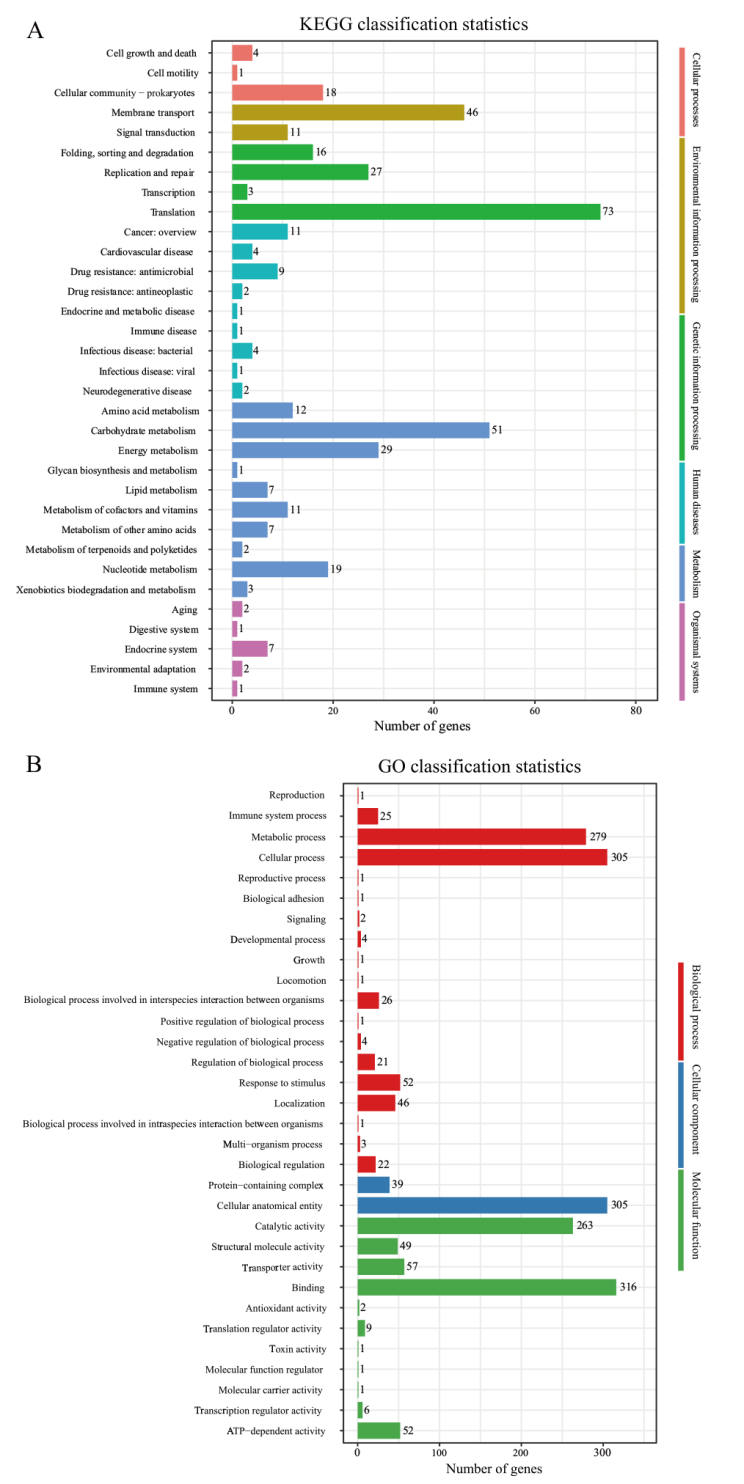
Fig. 8 Classification statistics diagrams of KEGG and GO annotation A: Classification statistics diagram of KEGG annotation. B: Classification statistics diagram of GO annotation
| 抗生素种类 Drug class | ARO登记号ARO accession | ARO名 ARO name |
|---|---|---|
| Aminoglycoside antibiotic | ARO:3005091 | RanA |
| Fluoroquinolone antibiotic | ARO:3003949 | efrB |
| Lincosamide antibiotic | ARO:3002882 | lmrD |
| ARO:3003746 | optrA | |
| ARO:3002825 | ErmY | |
| Macrolide antibiotic | ARO:3003949 | efrB |
| ARO:3003746 | optrA | |
| ARO:3002825 | ErmY | |
| ARO:3000535 | macB | |
| Mupirocin | ARO:3000510 | Staphylococcus aureus mupB conferring resistance to mupirocin |
| ARO:3000521 | Staphylococcus aureus mupA conferring resistance to mupirocin | |
| Nitroimidazole antibiotic | ARO:3003950 | msbA |
| Peptide antibiotic | ARO:3002987 | bcrA |
| ARO:3000501 | rpoB2 | |
| Tetracycline antibiotic | ARO:3004032 | tetA(46) |
| ARO:3000193 | tetT | |
| ARO:3002891 | otr(A) | |
| ARO:3003980 | tetA(58) | |
| ARO:3000556 | tet(44) | |
| ARO:3003980 | tetA(58) | |
| ARO:3003746 | optrA | |
| Rifamycin antibiotic | ARO:3003949 | efrB |
| ARO:3002825 | ErmY | |
| ARO:3000501 | rpoB2 | |
| Oxazolidinone antibiotic | ARO:3003746 | optrA |
| Pleuromutilin antibiotic | ARO:3003746 | optrA |
| Streptogramin antibiotic | ARO:3003746 | optrA |
| phenicol antibiotic | ARO:3003746 | optrA |
Table 2 Annotation of drug resistance genes
| 抗生素种类 Drug class | ARO登记号ARO accession | ARO名 ARO name |
|---|---|---|
| Aminoglycoside antibiotic | ARO:3005091 | RanA |
| Fluoroquinolone antibiotic | ARO:3003949 | efrB |
| Lincosamide antibiotic | ARO:3002882 | lmrD |
| ARO:3003746 | optrA | |
| ARO:3002825 | ErmY | |
| Macrolide antibiotic | ARO:3003949 | efrB |
| ARO:3003746 | optrA | |
| ARO:3002825 | ErmY | |
| ARO:3000535 | macB | |
| Mupirocin | ARO:3000510 | Staphylococcus aureus mupB conferring resistance to mupirocin |
| ARO:3000521 | Staphylococcus aureus mupA conferring resistance to mupirocin | |
| Nitroimidazole antibiotic | ARO:3003950 | msbA |
| Peptide antibiotic | ARO:3002987 | bcrA |
| ARO:3000501 | rpoB2 | |
| Tetracycline antibiotic | ARO:3004032 | tetA(46) |
| ARO:3000193 | tetT | |
| ARO:3002891 | otr(A) | |
| ARO:3003980 | tetA(58) | |
| ARO:3000556 | tet(44) | |
| ARO:3003980 | tetA(58) | |
| ARO:3003746 | optrA | |
| Rifamycin antibiotic | ARO:3003949 | efrB |
| ARO:3002825 | ErmY | |
| ARO:3000501 | rpoB2 | |
| Oxazolidinone antibiotic | ARO:3003746 | optrA |
| Pleuromutilin antibiotic | ARO:3003746 | optrA |
| Streptogramin antibiotic | ARO:3003746 | optrA |
| phenicol antibiotic | ARO:3003746 | optrA |
| 毒力因子名称 VFDB name | 相关基因 Related genes |
|---|---|
| <alpha>-Hemolysin | hlyB |
| Acinetobactin | bauE |
| Alginate regulation | mucD |
| Capsule | cpsB、FTT 0793、rpe |
| Cereulide | cesC |
| ClpC | ClpC |
| Cya | cyaB |
| Cytolysin | cylB |
| Dot/Icm | lpg2936 |
| EF-Tu | tufA |
| ESX-1 | eccA1 |
| FbpABC | fbpC |
| FeoAB | feoB |
| Flagella | flhF |
| FupA | feoB |
| HitABC | hitC |
| HP-NAP | napA |
| IgA1 protease | zmpC |
| LOS | licA |
| LplA1 | lplA1 |
| MgtBC | mgtB |
| MsrAB | msrA/B(pilB) |
| P97/P102 paralog family | mhp107、mhp683、p102、p97、lppT、p216、mhp385 |
| T4SS effectors | CBU 1566 |
| TlyC | tlyC |
| TTSS | escN、yscN、pscN |
| Type 1 fimbriae | fimE |
Table 3 Annotation of virulence factors
| 毒力因子名称 VFDB name | 相关基因 Related genes |
|---|---|
| <alpha>-Hemolysin | hlyB |
| Acinetobactin | bauE |
| Alginate regulation | mucD |
| Capsule | cpsB、FTT 0793、rpe |
| Cereulide | cesC |
| ClpC | ClpC |
| Cya | cyaB |
| Cytolysin | cylB |
| Dot/Icm | lpg2936 |
| EF-Tu | tufA |
| ESX-1 | eccA1 |
| FbpABC | fbpC |
| FeoAB | feoB |
| Flagella | flhF |
| FupA | feoB |
| HitABC | hitC |
| HP-NAP | napA |
| IgA1 protease | zmpC |
| LOS | licA |
| LplA1 | lplA1 |
| MgtBC | mgtB |
| MsrAB | msrA/B(pilB) |
| P97/P102 paralog family | mhp107、mhp683、p102、p97、lppT、p216、mhp385 |
| T4SS effectors | CBU 1566 |
| TlyC | tlyC |
| TTSS | escN、yscN、pscN |
| Type 1 fimbriae | fimE |
| [1] | Maksimović Z, Rifatbegović M, Loria GR, et al. Mycoplasma ovipneumoniae: a most variable pathogen[J]. Pathogens, 2022, 11(12): 1477. |
| [2] | Li Y, Jiang Z, Xue D, et al. Mycoplasma ovipneumoniae induces sheep airway epithelial cell apoptosis through an ERK signalling-mediated mitochondria pathway[J]. BMC microbiology. 2016, 16(1):222. |
| [3] |
Carmichael LE, St George TD, Sullivan ND, et al. Isolation, propagation, and characterization studies of an ovine Mycoplasma responsible for proliferative interstitial pneumonia[J]. Cornell Vet, 1972, 62(4): 654-679.
pmid: 4672899 |
| [4] | Balish M, Bertaccini A, Blanchard A, et al. Recommended rejection of the names Malacoplasma gen. nov., Mesomycoplasma gen. nov., Metamycoplasma gen. nov., Metamycoplasmataceae fam. nov., Mycoplasmoidaceae fam. nov., Mycoplasmoidales ord. nov., Mycoplasmoides gen. nov., Mycoplasmopsis gen. nov.[Gupta, Sawnani, Adeolu, Alnajar and Oren 2018]and all proposed species comb. nov. placed therein[J]. Int J Syst Evol Microbiol, 2019, 69(11): 3650-3653. |
| [5] | Gupta RS, Oren A. Necessity and rationale for the proposed name changes in the classification of Mollicutes species. Reply to: ‘Recommended rejection of the names Malacoplasma gen. nov., Mesomycoplasma gen. nov., Metamycoplasma gen. nov., Metamycoplasmataceae fam. nov., Mycoplasmoidaceae fam. nov., Mycoplasmoidales ord. nov., Mycoplasmoides gen. nov., Mycoplasmopsis gen. nov.[Gupta, Sawnani, Adeolu, Alnajar and Oren 2018]and all proposed species comb. nov. placed therein’, by M. Balish et al.(Int J Syst Evol Microbiol, 2019;69: 3650-3653)[J]. Int J Syst Evol Microbiol, 2020, 70(2): 1431-1438. |
| [6] | Gupta RS, Sawnani S, Adeolu M, et al. Phylogenetic framework for the phylum Tenericutes based on genome sequence data: proposal for the creation of a new order Mycoplasmoidales ord. nov., containing two new families Mycoplasmoidaceae fam. nov. and Metamycoplasmataceae fam. nov. harbouring Eperythrozoon, Ureaplasma and five novel Genera[J]. Antonie Van Leeuwenhoek, 2018, 111(9): 1583-1630. |
| [7] | 乌志勇. 绵羊肺炎支原体SY株分离鉴定及HSP70蛋白基因的克隆与分析[D]. 呼和浩特: 内蒙古农业大学, 2023. |
| Wu ZY. Isolation and identification of SY strain of Mycoplasma ovi-pneumoniae and gene cloning and analysis of HSP70 protein[D]. Hohhot: Inner Mongolia Agricultural University, 2023. | |
| [8] | 谭星. 绵羊肺炎支原体诱导巨噬细胞分泌的细胞外囊泡Lgals3bp蛋白促炎作用研究[D]. 银川: 宁夏大学, 2022. |
| Tan X. Proinflammatory effect of Lgals3bp protein in extracellular vesicles secreted by macrophages induced by Mycoplasma ovipneumoniae[D]. Yinchuan: Ningxia University, 2022. | |
| [9] | Gautier-Bouchardon AV. Antimicrobial resistance in Mycoplasma spp[J]. Microbiol Spectr, 2018, 6(4). |
| [10] | Ma CX, Li M, Peng H, et al. Mesomycoplasma ovipneumoniae from goats with respiratory infection: pathogenic characteristics, population structure, and genomic features[J]. BMC Microbiol, 2023, 23(1): 220. |
| [11] |
Kolmogorov M, Bickhart DM, Behsaz B, et al. metaFlye: scalable long-read metagenome assembly using repeat graphs[J]. Nat Methods, 2020, 17(11): 1103-1110.
doi: 10.1038/s41592-020-00971-x pmid: 33020656 |
| [12] | Walker BJ, Abeel T, Shea T, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement[J]. PLoS One, 2014, 9(11): e112963. |
| [13] | O’Leary NA, Wright MW, Brister JR, et al. Reference sequence(RefSeq)database at NCBI: current status, taxonomic expansion, and functional annotation[J]. Nucleic Acids Res, 2016, 44(D1): D733-D745. |
| [14] |
Boutet E, Lieberherr D, Tognolli M, et al. UniProtKB/swiss-prot, the manually annotated section of the UniProt KnowledgeBase: how to use the entry view[J]. Methods Mol Biol, 2016, 1374: 23-54.
doi: 10.1007/978-1-4939-3167-5_2 pmid: 26519399 |
| [15] | Galperin MY, Wolf YI, Makarova KS, et al. COG database update: focus on microbial diversity, model organisms, and widespread pathogens[J]. Nucleic Acids Res, 2021, 49(D1): D274-D281. |
| [16] | Kanehisa M, Furumichi M, Sato Y, et al. KEGG for taxonomy-based analysis of pathways and genomes[J]. Nucleic Acids Res, 2023, 51(D1): D587-D592. |
| [17] | Consortium TGO. The Gene Ontology Resource: 20 years and still GOing strong[J]. Nucleic Acids Res, 2019, 47(D1): D330-D338. |
| [18] | Alcock BP, Huynh W, Chalil R, et al. CARD 2023: expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database[J]. Nucleic Acids Res, 2023, 51(D1): D690-D699. |
| [19] | Lombard V, Golaconda Ramulu H, Drula E, et al. The carbohydrate-active enzymes database(CAZy)in 2013[J]. Nucleic Acids Res, 2014, 42(Database issue): D490-D495. |
| [20] | Urban M, Cuzick A, Seager J, et al. PHI-base in 2022: a multi-species phenotype database for Pathogen-Host Interactions[J]. Nucleic Acids Res, 2022, 50(D1): D837-D847. |
| [21] | Chen LH, Zheng DD, Liu B, et al. VFDB 2016: hierarchical and refined dataset for big data analysis—10 years on[J]. Nucleic Acids Res, 2016, 44(D1): D694-D697. |
| [22] | Saier MH, Reddy VS, Moreno-Hagelsieb G, et al. The Transporter Classification Database(TCDB): 2021 update[J]. Nucleic Acids Res, 2021, 49(D1): D461-D467. |
| [23] | Roberts RJ, Vincze T, Posfai J, et al. REBASE: a database for DNA restriction and modification: enzymes, genes and genomes[J]. Nucleic Acids Res, 2023, 51(D1): D629-D630. |
| [24] |
Nielsen H. Predicting secretory proteins with SignalP[J]. Methods Mol Biol, 2017, 1611: 59-73.
doi: 10.1007/978-1-4939-7015-5_6 pmid: 28451972 |
| [25] |
Wang YJ, Huang H, Sun MA, et al. T3DB: an integrated database for bacterial type III secretion system[J]. BMC Bioinformatics, 2012, 13: 66.
doi: 10.1186/1471-2105-13-66 pmid: 22545727 |
| [26] |
Krogh A, Larsson B, von Heijne G, et al. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes[J]. J Mol Biol, 2001, 305(3): 567-580.
doi: 10.1006/jmbi.2000.4315 pmid: 11152613 |
| [27] |
Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets[J]. Mol Biol Evol, 2016, 33(7): 1870-1874.
doi: 10.1093/molbev/msw054 pmid: 27004904 |
| [28] | Xie JM, Chen YR, Cai GJ, et al. Tree Visualization By One Table(tvBOT): a web application for visualizing, modifying and annotating phylogenetic trees[J]. Nucleic Acids Res, 2023, 51(W1): W587-W592. |
| [29] | Wang JD, Liu HY, Raheem A, et al. Exploring Mycoplasma ovipneumoniae NXNK2203 infection in sheep: insights from histopathology and whole genome sequencing[J]. BMC Vet Res, 2024, 20(1): 20. |
| [30] | Wang XH, Huang HB, Cheng C, et al. Complete genome sequence of Mycoplasma ovipneumoniae strain NM2010, which was isolated from a sheep in China[J]. J Integr Agric, 2014, 13(11): 2562-2563. |
| [31] | 韩瑞鑫. 绵羊肺炎支原体检测方法的建立及全基因组生物信息学分析[D]. 呼和浩特: 内蒙古农业大学, 2018. |
| Han RX. The establishment of detection method for Mycoplasma ovipneumoniae and analysis of it's genome bioinformatics[D]. Hohhot: Inner Mongolia Agricultural University, 2018. | |
| [32] | Yang FL, Tang C, Wang Y, et al. Genome sequence of Mycoplasma ovipneumoniae strain SC01[J]. J Bacteriol, 2011, 193(18): 5018. |
| [33] | 徐春光. 丝状支原体PG3株和绵羊肺炎支原体内蒙分离株基因组测序及可变表面脂蛋白基因克隆表达[D]. 呼和浩特: 内蒙古农业大学, 2013. |
| Xu CG. Genome sequencing of Mycoplasma mycoides Str.PG3and Mycoplasma ovipneumoniae from inner Mongolia and cloning and expression of variable surface lipoprotein genes[D]. Hohhot: Inner Mongolia Agricultural University, 2013. | |
| [34] | Hao HF, Maksimović Z, Ma LN, et al. Complete genome sequences of Mycoplasma ovipneumoniae strains 150 and 274, isolated from different regions in Bosnia and Herzegovina[J]. Microbiol Resour Announc, 2023, 12(3): e0001123. |
| [35] | Li M, Wang J, Hao XJ, et al. Complete Genome Sequence of Mycoplasma ovipneumoniae Strain Y98[C]. 哈尔滨: 第五届全国生物信息学与系统生物学学术大会论文集, 2012, 2:138-139. |
| Li M, Wang J, Hao XJ, et al. Complete Genome Sequence of Mycoplasma ovipneumoniae Strain Y98[C]. Harbin: Proceedings of the 5th National Conference on Bioinformatics and Systems Biology, 2012, 2:138-139. | |
| [36] |
Branton D, Deamer DW, Marziali A, et al. The potential and challenges of nanopore sequencing[J]. Nat Biotechnol, 2008, 26(10): 1146-1153.
doi: 10.1038/nbt.1495 pmid: 18846088 |
| [37] | 张金花. 绵羊肺炎支原体分离鉴定、病理组织学分析及靶器官主要细胞因子变化研究[D]. 银川: 宁夏大学, 2022. |
| Zhang JH. Isolation, identification and histopathological analysis of Mycoplasma ovipneumoniae and study on the change of main cytokines in target organs[D]. Yinchuan: Ningxia University, 2022. | |
| [38] | Liu W, Xiao SB, Li M, et al. Comparative genomic analyses of Mycoplasma hyopneumoniae pathogenic 168 strain and its high-passaged attenuated strain[J]. BMC Genomics, 2013, 14: 80. |
| [39] | 杨发龙, 汤承. 绵羊肺炎支原体毒力相关基因预测与分析[J]. 黑龙江畜牧兽医, 2013(19): 18-20. |
| Yang FL, Tang C. Prediction and analysis of the virulence- related genes in sheep Mycoplasma pneumoniae[J]. Heilongjiang Anim Sci Vet Med, 2013(19): 18-20. | |
| [40] | Mohamed AIS. 牛支原体HB0801株具有免疫原性分泌蛋白的鉴定[D]. 武汉: 华中农业大学, 2019. |
| Mohamed AIS. Identification of immunogenic secretory proteins of mycoplasma bovis HB0801[D]. Wuhan: Huazhong Agricultural University, 2019. |
| [1] | CHEN Bao-qiang, LI Ying-ying, MA Bo-ya, ROUZHAGULI Malike, YOULITUZI Naibi, SONG Jin-di, LIU Jun, WANG Xi-dong. Functional Analysis of the Type III Secreted Effector Gene aop2 in Acidovorax citrulli [J]. Biotechnology Bulletin, 2023, 39(6): 286-297. |
| [2] | WU Li-dan, RAN Xue-qin, NIU Xi, HUANG Shi-hui, LI Sheng, WANG Jia-fu. Genome Comparison and Virulence Factor Analysis of Pathogenic Escherichia coli from Porcine [J]. Biotechnology Bulletin, 2023, 39(12): 287-299. |
| [3] | LIU Li-hui, CHU Jin-hua, SUI Yu-xin, CHEN Yang, CHENG Gu-yue. Research Progress of Main Virulence Factors in Salmonella [J]. Biotechnology Bulletin, 2022, 38(9): 72-83. |
| [4] | XU Jun, WANG Rui, LI Rui, GAO Hai-xia, ZHANG Rui-liang, WANG Wen-jing, ZHAO Xia, LI Lin. RNA-seq Analysis of Drug-resistant Genes Associated with baeSR and acrB Double Gene Deletion Strains of Salmonella typhimurium [J]. Biotechnology Bulletin, 2019, 35(12): 76-84. |
| [5] | Ling Kong,Ding Shihua, Jin Juan,Wu Xingzhen. Detection of Pathogenic Aeromonas hydrophila in Andrias davidianus by Quadruple PCR [J]. Biotechnology Bulletin, 2014, 30(9): 201-207. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||