








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (8): 309-319.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0092
Previous Articles Next Articles
ZHANG A-na1( ), HAN Xue2,3, GU Tian-yi2,3, XIN Feng-jiao2,3(
), HAN Xue2,3, GU Tian-yi2,3, XIN Feng-jiao2,3( ), WANG Yu-lu2,3(
), WANG Yu-lu2,3( )
)
Received:2024-01-24
Online:2024-08-26
Published:2024-06-27
Contact:
XIN Feng-jiao, WANG Yu-lu
E-mail:zhangana123@163.com;2002hongzhi30@163.com;wnewyx@163.com
ZHANG A-na, HAN Xue, GU Tian-yi, XIN Feng-jiao, WANG Yu-lu. Preparation of Low-phenylalanine Casein by Novel Phenylalanine Ammonia-lyases Derived from Rhodotorula[J]. Biotechnology Bulletin, 2024, 40(8): 309-319.
| 蛋白名称 Protein name | 来源 Source | 基因号 Accession number | 氨基酸数目 Number of amino acids | 分子量 Molecular weight/kD | 等电点 pI |
|---|---|---|---|---|---|
| RmPAL | Rhodotorula mucilaginosa | P10248.2 | 713 | 76 | 6.72 |
| RdPAL | Rhodotorula diobovata | TNY17356.1 | 720 | 77 | 6.64 |
Table 1 Basic properties of RmPAL and RdPAL
| 蛋白名称 Protein name | 来源 Source | 基因号 Accession number | 氨基酸数目 Number of amino acids | 分子量 Molecular weight/kD | 等电点 pI |
|---|---|---|---|---|---|
| RmPAL | Rhodotorula mucilaginosa | P10248.2 | 713 | 76 | 6.72 |
| RdPAL | Rhodotorula diobovata | TNY17356.1 | 720 | 77 | 6.64 |
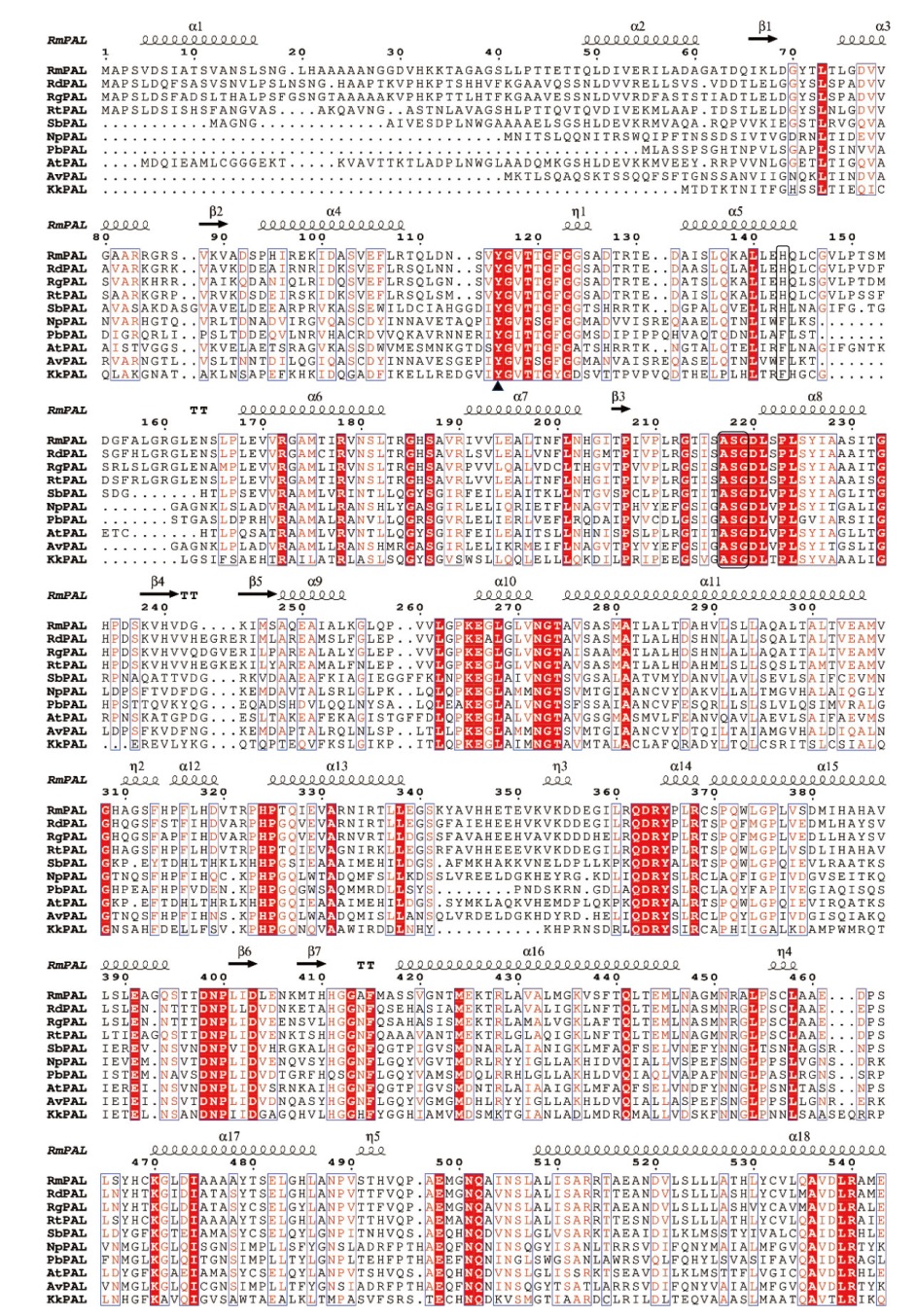
Fig. 2 Multiple sequence alignment of RmPAL and RdPAL The analysed PALs include: RgPAL(Rhodotorula glutinis, XP_018274290.1), RtPAL(Rhodotorula toruloides, XP_016272209.1), SbPAL(Sorghum bicolor, XP_002454198.1), NpPAL(Nostoc punctiforme, WP_012408693.1), PbPAL(Planctomyces brasiliensis, WP_150106093.1), AtPAL(Arabidopsis thaliana, NP_190894.1), AvPAL(Anabaena variabilis WP_011320679.1), KkPAL(Kangiella koreensis, WP_015781593.1). The catalytic amino acid Tyr is marked with a triangle. The MIO cofactor(Ala-Ser-Gly)and the substrate specific key amino acid His/Phe are marked with a black box
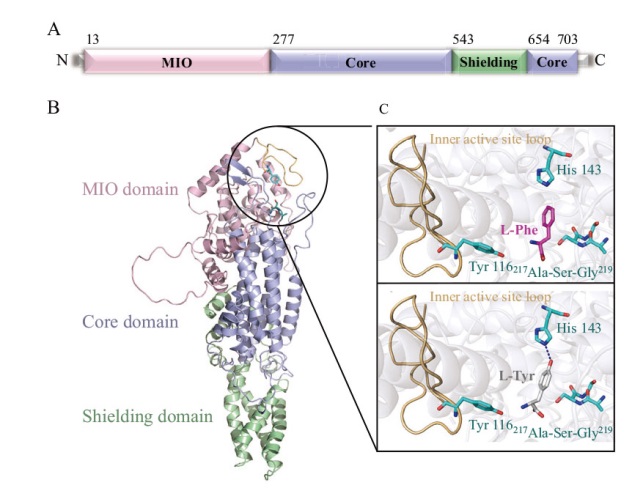
Fig. 4 Structure prediction of RmPAL A: Primary structure prediction of RmPAL; the MIO domain, core domain and shielding domain are colored by pink, cyan and green, respectively. B: AlphaFold2-based three-dimensional structure building of RmPAL, using the same coloring scheme as in A. C: Cartoon representation of L-Phe/L-Tyr binding to the active site of RmPAL. The catalytic amino acid Tyr 116, substrate switch His 143 and MIO(217 Ala-Ser-Gly 219)are depicted as green sticks. The inner active site loop is colored in wheat. The substrate L-Phe and L-Tyr are shown as pink and white sticks, respectively. Hydrogen bonding force between His 143 and L-Tyr is shown as a blue dashed line
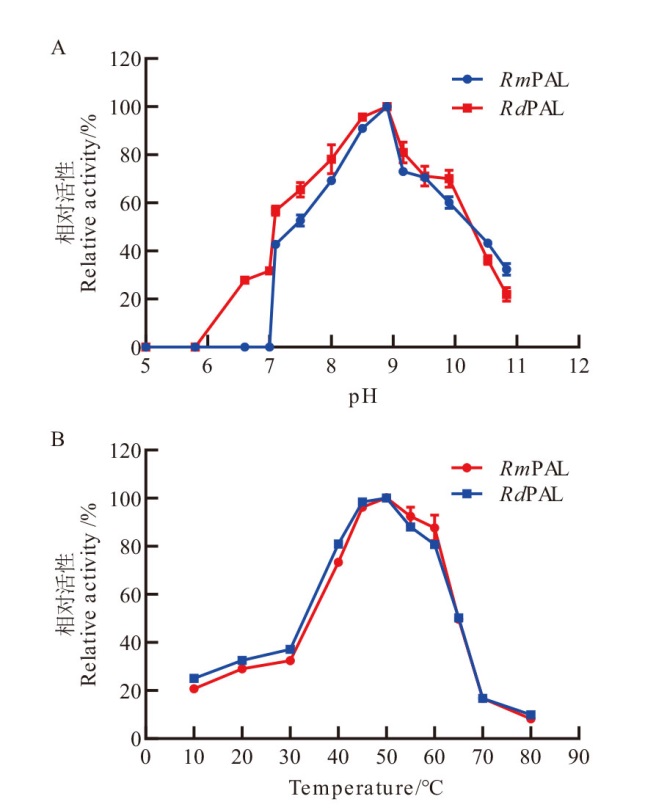
Fig. 6 Optimal conditions of RmPAL and RdPAL A: Effects of pH on the activities of RmPAL and RdPAL in the range of pH 5.0-11.0. B: Effects of temperature on the activities of RmPAL and RdPAL were determined at 10-80℃
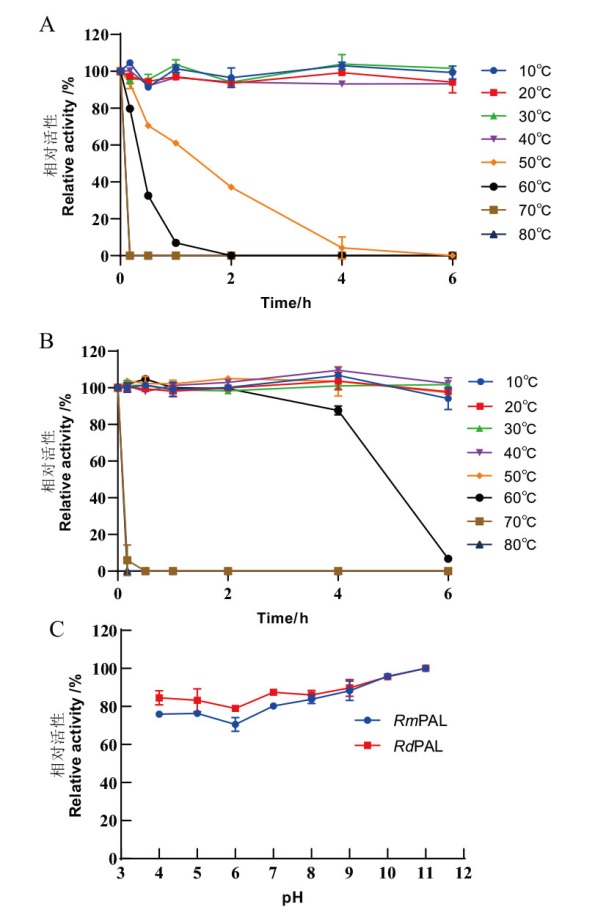
Fig. 7 Temperature and pH stability of RmPAL and Rd-PAL A, B: Thermal stability of RmPAL(A)and RdPAL(B)was determined after pre-incubated for 0-6 h at 10, 20, 30, 40, 50, 60, 70 and 80℃. C: The pH stability of RmPAL and RdPAL was determined after 1 h pretreatment in buffers of different pH values(pH 4.0-11.0)
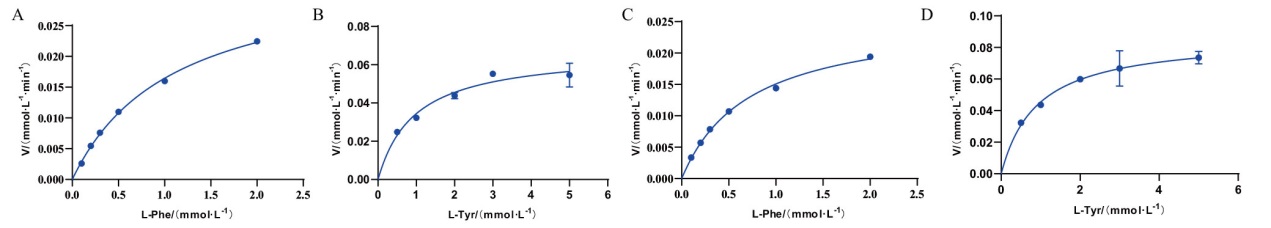
Fig. 8 Michaelis-Menten plots of RmPAL and RdPAL-catalyzed reactions Michaelis-Menten plots of RmPAL(A)and RdPAL(C)with L-Phe as the substrate. Michaelis-Menten plots of RmPAL(B)and RdPAL(D)with L-Tyr as the substrate.
| 蛋白名称Protein name | 底物Substate | Km/(mmol·L-1) | kcat/(s-1) | kcat/Km(mmol·L-1·s-1) | 倍数Fold | 参考文献Reference |
|---|---|---|---|---|---|---|
| RmPAL | L-Phe | 1.10±0.05 | 5.83±0.13 | 5.30 | 5 | This study |
| L-Tyr | 0.96±0.18 | 1.02±0.06 | 1.06 | This study | ||
| RdPAL | L-Phe | 0.72±0.04 | 4.47±0.11 | 6.21 | 4.7 | This study |
| L-Tyr | 0.91±0.16 | 1.20±0.07 | 1.32 | This study | ||
| RgPAL | L-Phe | 0.29 ± 0.01 | 0.22 ± 0.02 | 0.76 | 0.003 | [ |
| L-Tyr | 0.028± 0.002 | 7.12 ± 0.9 | 254 | [ | ||
| RtPAL | L-Phe | 0.54 ± 0.04 | 5.99 ± 0.06 | 11.03 | 2.24 | [ |
| L-Tyr | 0.21 ± 0.01 | 1.02 ± 0.01 | 4.92 | [ |
Table 2 Kinetic parameters of RmPAL and RdPAL in the hydrolysis of L-Phe and L-Tyr
| 蛋白名称Protein name | 底物Substate | Km/(mmol·L-1) | kcat/(s-1) | kcat/Km(mmol·L-1·s-1) | 倍数Fold | 参考文献Reference |
|---|---|---|---|---|---|---|
| RmPAL | L-Phe | 1.10±0.05 | 5.83±0.13 | 5.30 | 5 | This study |
| L-Tyr | 0.96±0.18 | 1.02±0.06 | 1.06 | This study | ||
| RdPAL | L-Phe | 0.72±0.04 | 4.47±0.11 | 6.21 | 4.7 | This study |
| L-Tyr | 0.91±0.16 | 1.20±0.07 | 1.32 | This study | ||
| RgPAL | L-Phe | 0.29 ± 0.01 | 0.22 ± 0.02 | 0.76 | 0.003 | [ |
| L-Tyr | 0.028± 0.002 | 7.12 ± 0.9 | 254 | [ | ||
| RtPAL | L-Phe | 0.54 ± 0.04 | 5.99 ± 0.06 | 11.03 | 2.24 | [ |
| L-Tyr | 0.21 ± 0.01 | 1.02 ± 0.01 | 4.92 | [ |
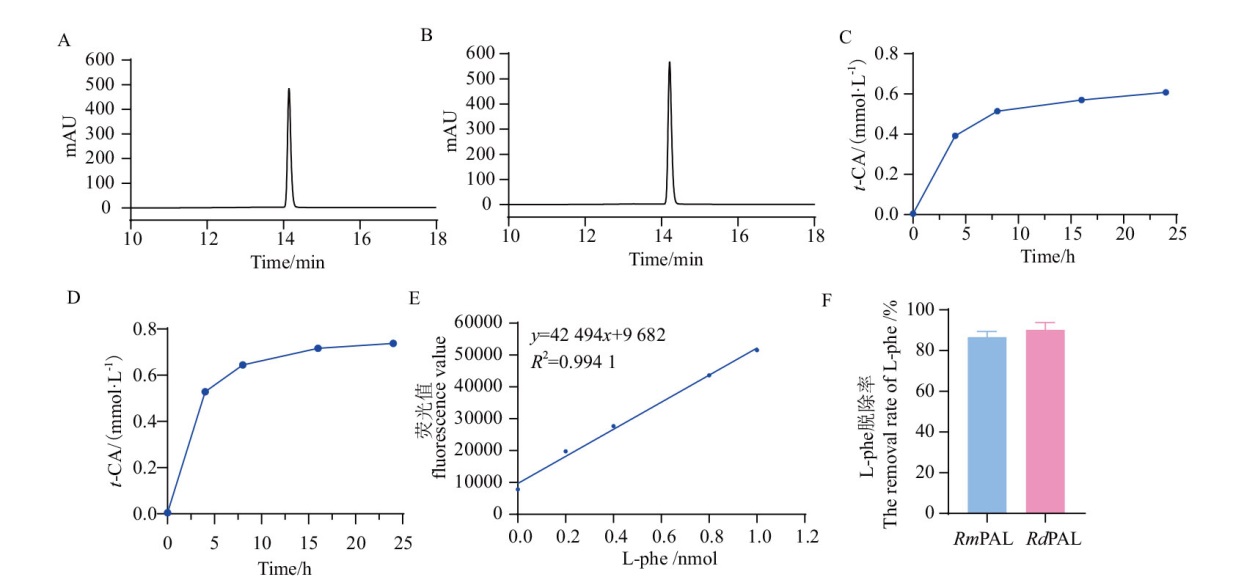
Fig. 9 HPLC analysis of the degradation of CAH by RmPAL and RdPAL A: Product t-CA of CAH hydrolysis by RmPAL for 24 h. B: Product t-CA of CAH hydrolysis by RdPAL for 24 h. C, D: The time dependence absorption profiles for the amount of product t-CA in CAH hydrolysis by RmPAL(C)and RdPAL(D). E: The standard curve of L-Phe. F: The removal rate of L-Phe after 24 h hydrolysis of CAH by RmPAL and RdPAL
| [1] |
van Spronsen FJ, Blau N, Harding C, et al. Phenylketonuria[J]. Nat Rev Dis Primers, 2021, 7: 36.
doi: 10.1038/s41572-021-00267-0 pmid: 34017006 |
| [2] | Levy HL, Sarkissian CN, Scriver CR. Phenylalanine ammonia lyase(PAL): from discovery to enzyme substitution therapy for phenylketonuria[J]. Mol Genet Metab, 2018, 124(4): 223-229. |
| [3] | Lu CH, Feng YW, He YX, et al. Foods for aromatic amino acid metabolism disorder: a review of current status, challenges and opportunities[J]. Food Rev Int, 2023, 39(9): 6630-6647. |
| [4] | van Spronsen FJ, van Wegberg AM, Ahring K, et al. Key European guidelines for the diagnosis and management of patients with phenylketonuria[J]. Lancet Diabetes Endocrinol, 2017, 5(9): 743-756. |
| [5] |
Lichter-Konecki U, Vockley J. Phenylketonuria: current treatments and future developments[J]. Drugs, 2019, 79(5): 495-500.
doi: 10.1007/s40265-019-01079-z pmid: 30864096 |
| [6] | Wiedemann A, Oussalah A, Jeannesson É, et al. Phenylketonuria, from diet to gene therapy[J]. Med Sci, 2020, 36(8/9): 725-734. |
| [7] |
盛晓静, 王强, 石爱民, 等. 制备低苯丙氨酸特膳食品的研究进展[J]. 食品科学, 2016, 37(21): 285-290.
doi: 10.7506/spkx1002-6630-201621048 |
| Sheng XJ, Wang Q, Shi AM, et al. Progress in the preparation of low-phenylalanine foods for special dietary use[J]. Food Sci, 2016, 37(21): 285-290. | |
| [8] | 周志伟, 张嘉芷, 许永红, 等. 低苯丙肽配方食品的研制[J]. 食品工业科技, 1999, 20(S1): 104-108. |
| Zhou ZW, Zhang JZ, Xu YH, et al. Development of low phenylpropanoid peptide formula food[J]. Sci Technol Food Ind, 1999, 20(S1): 104-108. | |
| [9] | Cabrera-Padilla RY, Pinto GA, Giordano RLC, et al. A new conception of enzymatic membrane reactor for the production of whey hydrolysates with low contents of phenylalanine[J]. Process Biochem, 2009, 44(3): 269-276. |
| [10] | Lopes DCF, Bizzotto CS, Carreira RL, et al. Removal of phenylalanine from protein hydrolysates prepared with rice[J]. Journal of Food Technology, 2008, 6(2): 57-65 |
| [11] |
Hyun MW, Yun YH, Kim JY, et al. Fungal and plant phenylalanine ammonia-lyase[J]. Mycobiology, 2011, 39(4): 257-265.
doi: 10.5941/MYCO.2011.39.4.257 pmid: 22783113 |
| [12] |
Cui JD, Qiu JQ, Fan XW, et al. Biotechnological production and applications of microbial phenylalanine ammonia lyase: a recent review[J]. Crit Rev Biotechnol, 2014, 34(3): 258-268.
doi: 10.3109/07388551.2013.791660 pmid: 23688066 |
| [13] | Ahmad R, Sami N, Perveen G, et al. Biochemical characterization of novel phenylalanine ammonia-lyase from Spirulina CPCC-695[J]. Protein J, 2022, 41(3): 414-423. |
| [14] | Fan SP, Chen W, Wei JC, et al. Molecular cloning and characterization of three phenylalanine ammonia-lyase genes from Schisandra chinensis[J]. Chin J Nat Med, 2022, 20(7): 527-536. |
| [15] | Koukol J, Conn EE. The metabolism of aromatic compounds in higher plants. IV. Purification and properties of the phenylalanine deaminase of Hordeum vulgare[J]. J Biol Chem, 1961, 236: 2692-2698. |
| [16] |
Zhang F, Ren J, Zhan JX. Identification and characterization of an efficient phenylalanine ammonia-lyase from Photorhabdus luminescens[J]. Appl Biochem Biotechnol, 2021, 193(4): 1099-1115.
doi: 10.1007/s12010-020-03477-6 pmid: 33411135 |
| [17] |
Moffitt MC, Louie GV, Bowman ME, et al. Discovery of two cyanobacterial phenylalanine ammonia lyases: kinetic and structural characterization[J]. Biochemistry, 2007, 46(4): 1004-1012.
doi: 10.1021/bi061774g pmid: 17240984 |
| [18] | Zhu LB, Cui WJ, Fang YQ, et al. Cloning, expression and characterization of phenylalanine ammonia-lyase from Rhodotorula glutinis[J]. Biotechnol Lett, 2013, 35(5): 751-756. |
| [19] |
Kawatra A, Dhankhar R, Mohanty A, et al. Biomedical applications of microbial phenylalanine ammonia lyase: current status and future prospects[J]. Biochimie, 2020, 177: 142-152.
doi: S0300-9084(20)30195-4 pmid: 32828824 |
| [20] | Castañeda MT, Adachi O, Hours RA. Reduction of L-phenylalanine in protein hydrolysates using L-phenylalanine ammonia-lyase from Rhodosporidium toruloides[J]. J Ind Microbiol Biotechnol, 2015, 42(10): 1299-1307. |
| [21] | Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server[J]. Nucleic Acids Res, 2014, 42(Web Server issue): W320-W324. |
| [22] |
Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11[J]. Mol Biol Evol, 2021, 38(7): 3022-3027.
doi: 10.1093/molbev/msab120 pmid: 33892491 |
| [23] |
Teufel F, Almagro Armenteros JJ, Johansen AR, et al. SignalP 6.0 predicts all five types of signal peptides using protein language models[J]. Nat Biotechnol, 2022, 40(7): 1023-1025.
doi: 10.1038/s41587-021-01156-3 pmid: 34980915 |
| [24] | Meena K, Alam S, Dalei SK, et al. Continuous preparation of low-phenylalanine formulations by treatment of edible protein with immobilized phenylalanine ammonia lyase[J]. Process Biochem, 2023, 131: 67-76. |
| [25] |
Dreßen A, Hilberath T, Mackfeld U, et al. Phenylalanine ammonia lyase from Arabidopsis thaliana(AtPAL2): a potent MIO-enzyme for the synthesis of non-canonical aromatic alpha-amino acids: part I: Comparative characterization to the enzymes from Petroselinum crispum(PcPAL1)and Rhodosporidium toruloides(RtPAL)[J]. J Biotechnol, 2017, 258: 148-157.
doi: S0168-1656(17)30159-1 pmid: 28392421 |
| [26] | Bu TT, Zhou MJ, Zheng JX, et al. Preparation and characterization of a low-phenylalanine whey hydrolysate using two-step enzymatic hydrolysis and macroporous resin adsorption[J]. LWT, 2020, 132: 109753. |
| [27] |
Babich OO, Pokrovsky VS, Anisimova NY, et al. Recombinant l-phenylalanine ammonia lyase from Rhodosporidium toruloides as a potential anticancer agent[J]. Biotechnol Appl Biochem, 2013, 60(3): 316-322.
doi: 10.1002/bab.1089 pmid: 23718781 |
| [28] | Varga A, Csuka P, Sonesouphap O, et al. A novel phenylalanine ammonia-lyase from Pseudozyma antarctica for stereoselective biotransformations of unnatural amino acids[J]. Catal Today, 2021, 366: 185-194. |
| [29] | Bata Z, Molnár Z, Madaras E, et al. Substrate tunnel engineering aided by X-ray crystallography and functional dynamics swaps the function of MIO-enzymes[J]. ACS Catal, 2021, 11(8): 4538-4549. |
| [30] | Brack Y, Sun CH, Yi D, et al. Exploring the substrate switch motif of aromatic ammonia lyases[J]. ChemBioChem, 2023, 24(23): e202300584. |
| [1] | QIAO Ye, ZHANG Nan, YANG Jian-hua, ZHANG Cui-ying, ZHU Lei-lei. Identification and Enzymatic Characterization of a Sugar Phosphatase [J]. Biotechnology Bulletin, 2024, 40(7): 299-306. |
| [2] | ZHANG Chen, ZHANG Tong-tong, LIU Hai-ping. Screening and Identification of Ethylene-forming Enzymes with High Activity and Thermostability [J]. Biotechnology Bulletin, 2022, 38(11): 269-276. |
| [3] | ZHANG Xue, TAN Yu-meng, JIANG Hai-xia, YANG Guang-yu. Directed Evolution of α-1,2-fucosyltransferase by a Single-cell Ultra-high-throughput Screening Method [J]. Biotechnology Bulletin, 2022, 38(1): 289-298. |
| [4] | HAO Xiang-yang, LIU Fan, WU Huan, WANG Bin, SUN Xue-li, XIANG Lei-lei, WANG Tian-chi, LAI Zhong-xiong, CHENG Chun-zhen. Cloning and Expression Analysis of GjPAL Genes in Gerbera jamesonni [J]. Biotechnology Bulletin, 2021, 37(6): 13-23. |
| [5] | SUN Xi-lin, JIANG Zhen-yan, LIU Zhi-yi, DAI Lu, SUN Fei, HUANG Wei. Improvement of Thermal Stability of Ganoderma lucidum Protein LZ-8 by Site-directed Mutation of Amino Acids [J]. Biotechnology Bulletin, 2020, 36(1): 23-28. |
| [6] | HUA Chen, LI Xin-xin, TU Tao, YANG Hong, LUO Hui-ying, CHEN Jia-ming, YAO Bin, BAI Ying-guo, PENG Shu-chuan. Improving the Thermal Stability of Lactate Oxidase by ETSS [J]. Biotechnology Bulletin, 2018, 34(8): 144-150. |
| [7] | MA Bo, ZHANG Ting-ting, WU Bao-xiang. Identification of Filamentous Fungus Strain DWL-C010 and Thermal Stability Analysis of Its Soluble Pigment [J]. Biotechnology Bulletin, 2018, 34(4): 186-193. |
| [8] | ZOU Tong-lei,WANG Fang-jun,HOU Sai-nan,SUN Xue,XU Nian-jun. On the Activities of Two Enzymes Related to Salicylic Acid Metabolism in Gracilariopsis lemaneiformis Under Adverse Environment [J]. Biotechnology Bulletin, 2016, 32(5): 194-199. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||