








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (6): 229-242.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0005
AN Chang1( ), XU Wen-bo2, LU Lin1, LI Deng-lin1, YAO Yi-xin3, LIN Yan-xiang4, YANG Cheng-zi4, QIN Yuan1(
), XU Wen-bo2, LU Lin1, LI Deng-lin1, YAO Yi-xin3, LIN Yan-xiang4, YANG Cheng-zi4, QIN Yuan1( ), ZHENG Ping1(
), ZHENG Ping1( )
)
Received:2025-01-03
Online:2025-06-26
Published:2025-06-30
Contact:
QIN Yuan, ZHENG Ping
E-mail:ancher0928@163.com;yuanqin@fafu.edu.cn;zhengping13@mails.ucas.ac.cn
AN Chang, XU Wen-bo, LU Lin, LI Deng-lin, YAO Yi-xin, LIN Yan-xiang, YANG Cheng-zi, QIN Yuan, ZHENG Ping. Comparative Analysis of Glehnia littoralis from Different Geographic Regions Based on the Characteristics of Chloroplast Genome[J]. Biotechnology Bulletin, 2025, 41(6): 229-242.
| 类别 Category | 分组 Gene group | 基因 Gene |
|---|---|---|
光合作用 Photosynthesis | 光系统I亚基 Subunits of photosystem I | psaA, psaB, psaC, psaI, psaJ |
光系统II亚基 Subunits of photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | |
NADH脱氢酶亚基 Subunits of NADH dehydrogenase | ndhA*, ndhB*(2), ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | |
细胞色素b/f 复合体亚基 Subunits of cytochrome b/f complex | petA, petB*, petD*, petG, petL, petN | |
ATP合酶亚基 Subunits of ATP synthase | atpA, atpB, atpE, atpF*, atpH, atpI | |
Rubisco大亚基 Large subunit of rubisco | rbcL | |
自我复制 Self-replication | 大核糖体亚基蛋白 Proteins of large ribosomal subunit | rpl14, rpl16*, rpl2*, rpl20, rpl22, rpl23, rpl32, rpl33, rpl36 |
小核糖体亚基蛋白 Proteins of small ribosomal subunit | rps11, rps12**(2), rps14, rps15, rps16*, rps18, rps19, rps2, rps3, rps4, rps7(2), rps8 | |
RNA聚合酶的亚基 Subunits of RNA polymerase | rpoA, rpoB, rpoC1*, rpoC2 | |
核糖体RNA Ribosomal RNAs | rrn16(2), rrn23(2), rrn4.5(2), rrn5(2) | |
转运RNA Transfer RNAs | trnA-UGC*(2), trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnG-GCC, trnG-UCC*, trnH-GUG, trnI-CAU, trnI-GAU*(2), trnK-UUU*, trnL-CAA(2), trnL-UAA*, trnL-UAG, trnM-CAU, trnN-GUU(2), trnP-UGG, trnQ-UUG, trnR-ACG(2), trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC(2), trnV-UAC*, trnW-CCA, trnY-GUA, trnfM-CAU | |
其他基因 Other genes | 成熟酶 Maturase | matK |
蛋白酶 Protease | clpP** | |
包膜膜蛋白 Envelope membrane protein | cemA | |
乙酰辅酶A羧化酶 Acetyl-CoA carboxylase | accD | |
c型细胞色素合成基因 c-type cytochrome synthesis gene | ccsA | |
翻译起始因子 Translation initiation factor | infA | |
功能未知 Genes of unknown function | 保守的假定叶绿体开放阅读框ORF Conserved hypothetical chloroplast ORF | ycf1(2), ycf15(2), ycf2, ycf3**, ycf4 |
Table 1 Functional annotation and classification of genes in the chloroplast genome of G. littoralis
| 类别 Category | 分组 Gene group | 基因 Gene |
|---|---|---|
光合作用 Photosynthesis | 光系统I亚基 Subunits of photosystem I | psaA, psaB, psaC, psaI, psaJ |
光系统II亚基 Subunits of photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | |
NADH脱氢酶亚基 Subunits of NADH dehydrogenase | ndhA*, ndhB*(2), ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | |
细胞色素b/f 复合体亚基 Subunits of cytochrome b/f complex | petA, petB*, petD*, petG, petL, petN | |
ATP合酶亚基 Subunits of ATP synthase | atpA, atpB, atpE, atpF*, atpH, atpI | |
Rubisco大亚基 Large subunit of rubisco | rbcL | |
自我复制 Self-replication | 大核糖体亚基蛋白 Proteins of large ribosomal subunit | rpl14, rpl16*, rpl2*, rpl20, rpl22, rpl23, rpl32, rpl33, rpl36 |
小核糖体亚基蛋白 Proteins of small ribosomal subunit | rps11, rps12**(2), rps14, rps15, rps16*, rps18, rps19, rps2, rps3, rps4, rps7(2), rps8 | |
RNA聚合酶的亚基 Subunits of RNA polymerase | rpoA, rpoB, rpoC1*, rpoC2 | |
核糖体RNA Ribosomal RNAs | rrn16(2), rrn23(2), rrn4.5(2), rrn5(2) | |
转运RNA Transfer RNAs | trnA-UGC*(2), trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnG-GCC, trnG-UCC*, trnH-GUG, trnI-CAU, trnI-GAU*(2), trnK-UUU*, trnL-CAA(2), trnL-UAA*, trnL-UAG, trnM-CAU, trnN-GUU(2), trnP-UGG, trnQ-UUG, trnR-ACG(2), trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC(2), trnV-UAC*, trnW-CCA, trnY-GUA, trnfM-CAU | |
其他基因 Other genes | 成熟酶 Maturase | matK |
蛋白酶 Protease | clpP** | |
包膜膜蛋白 Envelope membrane protein | cemA | |
乙酰辅酶A羧化酶 Acetyl-CoA carboxylase | accD | |
c型细胞色素合成基因 c-type cytochrome synthesis gene | ccsA | |
翻译起始因子 Translation initiation factor | infA | |
功能未知 Genes of unknown function | 保守的假定叶绿体开放阅读框ORF Conserved hypothetical chloroplast ORF | ycf1(2), ycf15(2), ycf2, ycf3**, ycf4 |
登录号 GenBank ID | 产地 Distribution | 大小 Size (bp) | 蛋白编码基因 PCG | 转运RNA tRNA | 核糖体RNA rRNA | 基因数 Genes | GC (%) | 长度 Length (bp) | ||
|---|---|---|---|---|---|---|---|---|---|---|
| LSC | SSC | IRb/a | ||||||||
| KT153022.1 | 韩国1 | 147 467 | 85 | 36 | 8 | 129 | 37.51 | 93 493 | 17 546 | 18 214 |
| NC_034645.1 | 韩国2 | 147 477 | 85 | 36 | 8 | 129 | 37.51 | 93 496 | 17 555 | 18 213 |
| OR004233.1 | 中国台湾 | 147 445 | 85 | 36 | 8 | 129 | 37.52 | 92 373 | 17 548 | 18 762 |
| PQ563254.1 | 福建 | 147 493 | 85 | 36 | 8 | 129 | 37.51 | 93 218 | 17 545 | 18 365 |
| MH142518.1 | 深圳 | 147 552 | 85 | 36 | 8 | 129 | 37.51 | 93 277 | 17 545 | 18 365 |
| OQ863734.1 | 浙江 | 147 507 | 85 | 36 | 8 | 129 | 37.51 | 93 231 | 17 546 | 18 365 |
Table 2 Characteristics of chloroplast genomes of G. littoralis from 6 different geographic distributions
登录号 GenBank ID | 产地 Distribution | 大小 Size (bp) | 蛋白编码基因 PCG | 转运RNA tRNA | 核糖体RNA rRNA | 基因数 Genes | GC (%) | 长度 Length (bp) | ||
|---|---|---|---|---|---|---|---|---|---|---|
| LSC | SSC | IRb/a | ||||||||
| KT153022.1 | 韩国1 | 147 467 | 85 | 36 | 8 | 129 | 37.51 | 93 493 | 17 546 | 18 214 |
| NC_034645.1 | 韩国2 | 147 477 | 85 | 36 | 8 | 129 | 37.51 | 93 496 | 17 555 | 18 213 |
| OR004233.1 | 中国台湾 | 147 445 | 85 | 36 | 8 | 129 | 37.52 | 92 373 | 17 548 | 18 762 |
| PQ563254.1 | 福建 | 147 493 | 85 | 36 | 8 | 129 | 37.51 | 93 218 | 17 545 | 18 365 |
| MH142518.1 | 深圳 | 147 552 | 85 | 36 | 8 | 129 | 37.51 | 93 277 | 17 545 | 18 365 |
| OQ863734.1 | 浙江 | 147 507 | 85 | 36 | 8 | 129 | 37.51 | 93 231 | 17 546 | 18 365 |
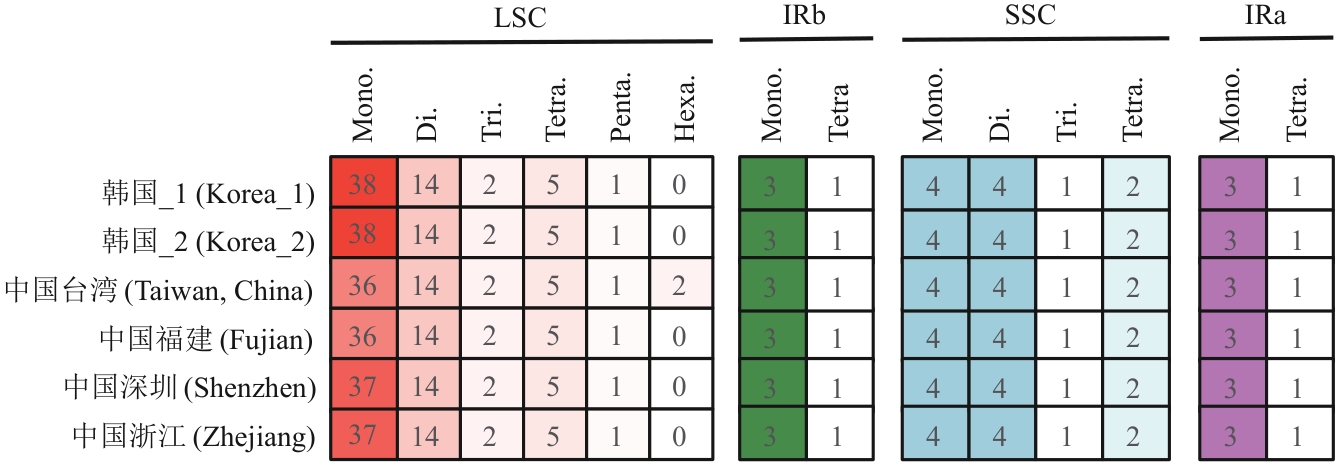
Fig. 2 Comparison of SSRs in the chloroplast genomes of G. littoralis from 6 different geographic distributionsMono: Mononucleotide repeat. Di: Dinucleotide repeat. Tri: Trinucleotide repeat. Tera: Tetranucleotide repeat. Penta: Pentanucleotide repeat. Hexa: Hexanucleotide repeat
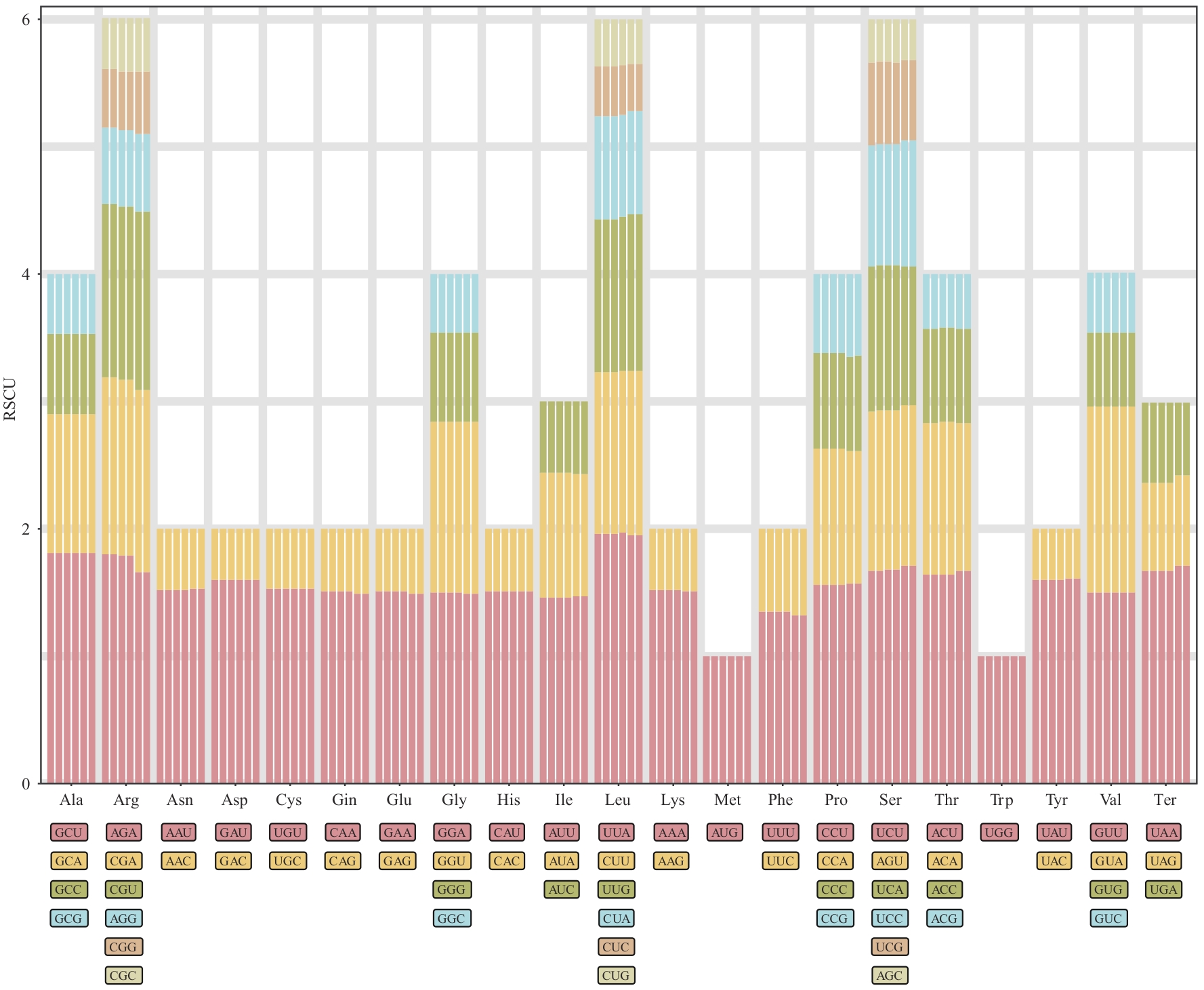
Fig. 3 Relative synonymous codon usage in the chloroplast genomes of G. littoralis from 6 different geographic distributionsFrom left to right, the geographic regions are: Korea_1, Korea_2, Taiwan (China), Fujian, Shenzhen, and Zhejiang
氨基酸 Amino acid | 密码子 Codon | 数量 Count | 同义密码子频率 RSCU | 氨基酸 Amino acid | 密码子 Codon | 数量 Count | 同义密码子频率 RSCU |
|---|---|---|---|---|---|---|---|
| Ala | GCA | 303-312 | 1.09 | Leu | CTT | 426-457 | 1.27-1.29 |
| Ala | GCC | 175-180 | 0.63 | Leu | CTC | 124-140 | 0.37-0.39 |
| Ala | GCG | 129-133 | 0.46-0.47 | Leu | CTG | 117-133 | 0.35-0.37 |
| Ala | GCT | 504-515 | 1.81 | Lys | AAG | 216-266 | 0.48-0.49 |
| Arg | AGG | 113-123 | 0.6-0.61 | Lys | AAA | 670-835 | 1.51-1.52 |
| Arg | CGA | 260-284 | 1.38-1.4 | Met | ATG | 435-467 | 1.00 |
| Arg | AGA | 309-368 | 1.66-1.8 | Phe | TTT | 681-781 | 1.32-1.35 |
| Arg | CGC | 78-81 | 0.4-0.42 | Phe | TTC | 351-373 | 0.65-0.68 |
| Arg | CGG | 91-95 | 0.46-0.49 | Pro | CCT | 312-338 | 1.56-1.57 |
| Arg | CGT | 266-278 | 1.36-1.43 | Pro | CCG | 128-135 | 0.62-0.64 |
| Asn | AAC | 200-235 | 0.47-0.48 | Pro | CCA | 207-231 | 1.04-1.07 |
| Asn | AAT | 648-753 | 1.52-1.53 | Pro | CCC | 148-163 | 0.74-0.75 |
| Asp | GAT | 619-673 | 1.60 | Ser | AGC | 73-85 | 0.31-0.33 |
| Asp | GAC | 154-167 | 0.40 | Ser | AGT | 293-321 | 1.25-1.26 |
| Cys | TGT | 155-164 | 1.53 | Ser | TCT | 397-428 | 1.67-1.71 |
| Cys | TGC | 47-50 | 0.47 | Ser | TCG | 147-165 | 0.63-0.65 |
| Gin | CAA | 512-570 | 1.49-1.51 | Ser | TCA | 253-290 | 1.09-1.14 |
| Gin | CAG | 177-185 | 0.49-0.51 | Ser | TCC | 230-243 | 0.95-0.99 |
| Glu | GAA | 714-815 | 1.49-1.51 | Thr | ACC | 182-198 | 0.74 |
| Glu | GAG | 246-265 | 0.49-0.51 | Thr | ACA | 285-320 | 1.16-1.2 |
| Gly | GGT | 469-485 | 1.34-1.35 | Thr | ACG | 104-114 | 0.42-0.43 |
| Gly | GGG | 245-253 | 0.70 | Thr | ACT | 409-439 | 1.64-1.67 |
| Gly | GGA | 518-542 | 1.49-1.5 | Trp | TGG | 339-370 | 1.00 |
| Gly | GGC | 160-168 | 0.46 | Tyr | TAC | 139-159 | 0.39-0.4 |
| His | CAT | 353-380 | 1.51 | Tyr | TAT | 566-636 | 1.6-1.61 |
| His | CAC | 115-123 | 0.49 | Val | GTA | 398-418 | 1.46 |
| Ile | ATC | 298-327 | 0.56-0.57 | Val | GTC | 125-134 | 0.46-0.47 |
| Ile | ATA | 506-571 | 0.96-0.98 | Val | GTG | 159-167 | 0.58 |
| Ile | ATT | 773-850 | 1.46-1.47 | Val | GTT | 409-430 | 1.50 |
| Leu | TTG | 406-435 | 1.2-1.23 | Ter | TAG | 12-12 | 0.69-0.71 |
| Leu | TTA | 646-708 | 1.95-1.97 | Ter | TGA | 10-11 | 0.59-0.63 |
| Leu | CTA | 269-290 | 0.8-0.81 | Ter | TAA | 29-29 | 1.67-1.71 |
Table 3 Codon usage bias analysis of protein-coding sequences in 6 G. littoralis chloroplast genomes
氨基酸 Amino acid | 密码子 Codon | 数量 Count | 同义密码子频率 RSCU | 氨基酸 Amino acid | 密码子 Codon | 数量 Count | 同义密码子频率 RSCU |
|---|---|---|---|---|---|---|---|
| Ala | GCA | 303-312 | 1.09 | Leu | CTT | 426-457 | 1.27-1.29 |
| Ala | GCC | 175-180 | 0.63 | Leu | CTC | 124-140 | 0.37-0.39 |
| Ala | GCG | 129-133 | 0.46-0.47 | Leu | CTG | 117-133 | 0.35-0.37 |
| Ala | GCT | 504-515 | 1.81 | Lys | AAG | 216-266 | 0.48-0.49 |
| Arg | AGG | 113-123 | 0.6-0.61 | Lys | AAA | 670-835 | 1.51-1.52 |
| Arg | CGA | 260-284 | 1.38-1.4 | Met | ATG | 435-467 | 1.00 |
| Arg | AGA | 309-368 | 1.66-1.8 | Phe | TTT | 681-781 | 1.32-1.35 |
| Arg | CGC | 78-81 | 0.4-0.42 | Phe | TTC | 351-373 | 0.65-0.68 |
| Arg | CGG | 91-95 | 0.46-0.49 | Pro | CCT | 312-338 | 1.56-1.57 |
| Arg | CGT | 266-278 | 1.36-1.43 | Pro | CCG | 128-135 | 0.62-0.64 |
| Asn | AAC | 200-235 | 0.47-0.48 | Pro | CCA | 207-231 | 1.04-1.07 |
| Asn | AAT | 648-753 | 1.52-1.53 | Pro | CCC | 148-163 | 0.74-0.75 |
| Asp | GAT | 619-673 | 1.60 | Ser | AGC | 73-85 | 0.31-0.33 |
| Asp | GAC | 154-167 | 0.40 | Ser | AGT | 293-321 | 1.25-1.26 |
| Cys | TGT | 155-164 | 1.53 | Ser | TCT | 397-428 | 1.67-1.71 |
| Cys | TGC | 47-50 | 0.47 | Ser | TCG | 147-165 | 0.63-0.65 |
| Gin | CAA | 512-570 | 1.49-1.51 | Ser | TCA | 253-290 | 1.09-1.14 |
| Gin | CAG | 177-185 | 0.49-0.51 | Ser | TCC | 230-243 | 0.95-0.99 |
| Glu | GAA | 714-815 | 1.49-1.51 | Thr | ACC | 182-198 | 0.74 |
| Glu | GAG | 246-265 | 0.49-0.51 | Thr | ACA | 285-320 | 1.16-1.2 |
| Gly | GGT | 469-485 | 1.34-1.35 | Thr | ACG | 104-114 | 0.42-0.43 |
| Gly | GGG | 245-253 | 0.70 | Thr | ACT | 409-439 | 1.64-1.67 |
| Gly | GGA | 518-542 | 1.49-1.5 | Trp | TGG | 339-370 | 1.00 |
| Gly | GGC | 160-168 | 0.46 | Tyr | TAC | 139-159 | 0.39-0.4 |
| His | CAT | 353-380 | 1.51 | Tyr | TAT | 566-636 | 1.6-1.61 |
| His | CAC | 115-123 | 0.49 | Val | GTA | 398-418 | 1.46 |
| Ile | ATC | 298-327 | 0.56-0.57 | Val | GTC | 125-134 | 0.46-0.47 |
| Ile | ATA | 506-571 | 0.96-0.98 | Val | GTG | 159-167 | 0.58 |
| Ile | ATT | 773-850 | 1.46-1.47 | Val | GTT | 409-430 | 1.50 |
| Leu | TTG | 406-435 | 1.2-1.23 | Ter | TAG | 12-12 | 0.69-0.71 |
| Leu | TTA | 646-708 | 1.95-1.97 | Ter | TGA | 10-11 | 0.59-0.63 |
| Leu | CTA | 269-290 | 0.8-0.81 | Ter | TAA | 29-29 | 1.67-1.71 |
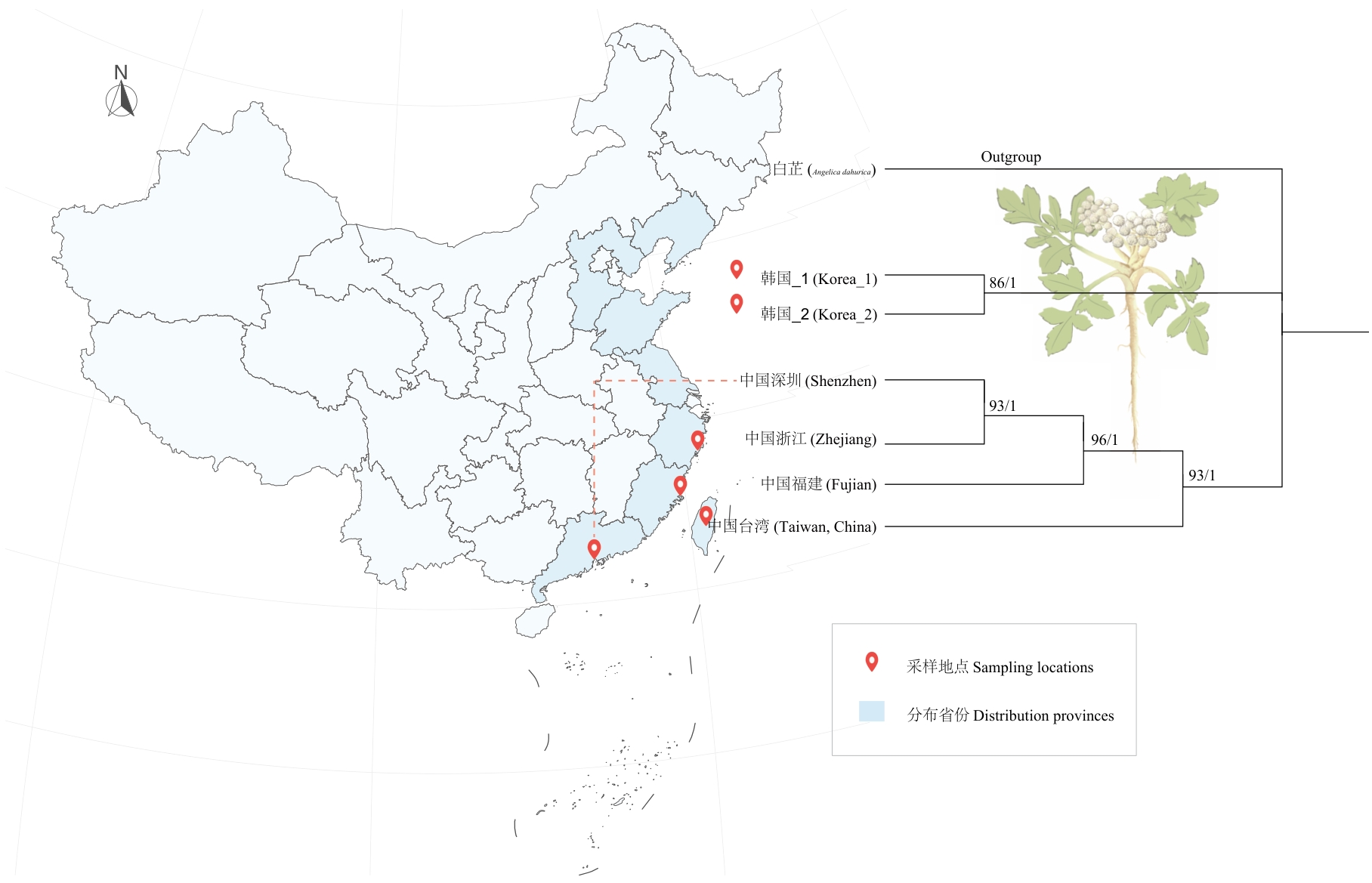
Fig. 6 Phylogenetic relationships of G. littoralis from 6 different geographic distributions inferred using Maximum Likelihood (ML) and Bayesian Inference (BI) methodsThe numbers on the branches indicate the support values from the ML method (BP) and the BI method (PP), respectively. The map of China is sourced from the Ministry of Natural Resources website (http://www.mnr.gov.cn), with the review number GS(2024) 0650
| 1 | 屠鹏飞, 冷青松, 徐国钧, 等. 莱阳参的生药鉴定 [J]. 中药材, 1999, 22(4): 174-176. |
| Tu PF, Leng QS, Xu GJ, et al. Pharmacognostical studies on Radix glehniae (Glehnia littoralis) [J]. J Chin Med Mater, 1999, 22(4): 174-176. | |
| 2 | 安莹, 张姗姗, 张云, 等. 北沙参化学成分、药理作用研究进展及质量标志物(Q-Marker)预测 [J]. 中草药, 2024, 55(2): 640-656. |
| An Y, Zhang SS, Zhang Y, et al. Research progress on chemical constituents, pharmacological effects and quality marker prediction of Glehniae radix [J]. Chin Tradit Herb Drugs, 2024, 55(2): 640-656. | |
| 3 | Li SY, Xu N, Fang QQ, et al. Glehnia littoralis Fr. schmidtex miq.: a systematic review on ethnopharmacology, chemical composition, pharmacology and quality control [J]. J Ethnopharmacol, 2023, 317: 116831. |
| 4 | Chen Y, Lei LJ, Bi YQ, et al. Quality control of glehniae Radix, the root of Glehnia littoralis Fr. Schmidt ex miq., along its value chains [J]. Front Pharmacol, 2021, 12: 729554. |
| 5 | 李宝国, 李军, 欧阳兵. 不同产地北沙参的氨基酸检测分析 [J]. 山东中医药大学学报, 2013, 37(5): 444-445. |
| Li BG, Li J, Ouyang B. Amino acid detection and analysis of radix glehniae from different sources [J]. J Shandong Univ Tradit Chin Med, 2013, 37(5): 444-445. | |
| 6 | 叶国华, 张钦德, 许一平, 等. 不同产地北沙参中重金属含量的测定 [J]. 中药新药与临床药理, 2013, 24(4): 407-410. |
| Ye GH, Zhang QD, Xu YP, et al. Determination of heavy metals in Radix glehniae from different producing areas [J]. Tradit Chin Drug Res Clin Pharmacol, 2013, 24(4): 407-410. | |
| 7 | 郑旭光, 陈钟, 项峰, 等. 河北道地药材北沙参HPLC - PDA指纹图谱研究 [J]. 药物分析杂志, 2011, 31(9): 1683-1688. |
| Zheng XG, Chen Z, Xiang F, et al. Study on HPLC-PDA fingerprints of Radix glehniae from Hebei Province [J]. Chin J Pharm Anal, 2011, 31(9): 1683-1688. | |
| 8 | 刘小芬, 赖冰, 宋秀碧, 等. HPLC法分析闽产北沙参3种香豆素含量 [J]. 中国中医药现代远程教育, 2020, 18(10): 107-110. |
| Liu XF, Lai B, Song XB, et al. Content analysis of three coumarins in glehniae Radix from Fujian Province by HPLC [J]. Chin Med Mod Distance Educ China, 2020, 18(10): 107-110. | |
| 9 | 张晴. 不同种源北沙参表型特征、香豆素类化合物含量及IPT基因表达差异研究 [D]. 烟台: 烟台大学, 2023. |
| Zhang Q. Differences in phenotypic identificarion、coumarin content and expression of key enzyme genes in Glehnia littoralis from different origins [D]. Yantai: Yantai University, 2023. | |
| 10 | 张敏, 杨洪晓. 滨海沙滩珊瑚菜种群的空间格局及其对岸垄的响应 [J]. 生态学杂志, 2015, 34(1): 47-52. |
| Zhang M, Yang HX. Spatial patterns of Glehnia littoralis population on sandy seashore and their responses to artificial beach ridge [J]. Chin J Ecol, 2015, 34(1): 47-52. | |
| 11 | 刘小芬, 张明孝, 陈勇, 等. 福建省长江澳珊瑚菜样地调查与群落特征 [J]. 中国野生植物资源, 2017, 36(6): 57-64. |
| Liu XF, Zhang MX, Chen Y, et al. Sample plot investigation and community characteristics of Glehnia littoralis in changjiang'ao bay, Fujian Province [J]. Chin Wild Plant Resour, 2017, 36(6): 57-64. | |
| 12 | An C, Ye KZ, Jiang RF, et al. Cytological analysis of flower development, insights into suitable growth area and genomic background: implications for Glehnia littoralis conservation and sustainable utilization [J]. BMC Plant Biol, 2024, 24(1): 895. |
| 13 | 陈梓媛, 华中一, 袁媛. 全球物种数量最多的叶绿体基因组数据库的建立及应用进展 [J]. 中国中药杂志, 2024, 49(23): 6257-6263. |
| Chen ZY, Hua ZY, Yuan Y. Establishment and application of chloroplast genome database with the largest number of species in world [J]. China J Chin Mater Med, 2024, 49(23): 6257-6263. | |
| 14 | 韩建萍, 宋经元, 姚辉, 等. 中药材DNA条形码鉴定的基因序列比较 [J]. 中国中药杂志, 2012, 37(8): 1056-1061. |
| Han JP, Song JY, Yao H, et al. Comparison of DNA barcoders in identifying medicinal materials [J]. China J Chin Mater Med, 2012, 37(8): 1056-1061. | |
| 15 | Chen SL, Yin XM, Han JP, et al. DNA barcoding in herbal medicine: Retrospective and prospective [J]. J Pharm Anal, 2023, 13(5): 431-441. |
| 16 | Lee SC, Lee HO, Kim K, et al. The complete chloroplast genome sequence of the medicinal plant Glehnia littoralis F.Schmidt ex Miq. (Apiaceae) [J]. Mitochondrial DNA A Part A, 2016, 27(5): 3674-3675. |
| 17 | Zhou YF, Geng ML, Li MM. The complete chloroplast genome of Glehnia littoralis, an Endangered medicinal herb of Apiaceae family [J]. Mitochondrial DNA B Resour, 2018, 3(2): 1013-1014. |
| 18 | 孙月琪, 李密密, 周义峰. 珊瑚菜叶绿体基因组密码子使用偏性分析 [J]. 植物资源与环境学报, 2023, 32(6): 1-10. |
| Sun YQ, Li MM, Zhou YF. Analysis on Codon usage bias of chloroplast genome of Glehnia littoralis [J]. J Plant Resour Environ, 2023, 32(6): 1-10. | |
| 19 | Li WW, Liu SL, Wang SM, et al. A single origin and high genetic diversity of cultivated medicinal herb Glehnia littoralis subsp. littoralis (Apiaceae) deciphered by SSR marker and phenotypic analysis [J]. PLoS One, 2024, 19(8): e0308369. |
| 20 | Li WW, Li B, Zhang P, et al. Potential biological mechanisms underlying the endangered status of Glehnia littoralis revealed by nrDNA ITS and RAPD analyses [J]. Biotechnol Biotechnol Equip, 2020, 34(1): 1243-1251. |
| 21 | Robbins EHJ, Kelly S. The evolutionary constraints on angiosperm chloroplast adaptation [J]. Genome Biol Evol, 2023, 15(6): evad101. |
| 22 | Daniell H, Lin CS, Yu M, et al. Chloroplast genomes: diversity, evolution, and applications in genetic engineering [J]. Genome Biol, 2016, 17(1): 134. |
| 23 | Jin JJ, Yu WB, Yang JB, et al. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes [J]. Genome Biol, 2020, 21(1): 241. |
| 24 | Tillich M, Lehwark P, Pellizzer T, et al. GeSeq - versatile and accurate annotation of organelle genomes [J]. Nucleic Acids Res, 2017, 45(W1): W6-W11. |
| 25 | Greiner S, Lehwark P, Bock R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes [J]. Nucleic Acids Res, 2019, 47(W1): W59-W64. |
| 26 | Huang LJ, Yu HX, Wang Z, et al. CPStools: a package for analyzing chloroplast genome sequences [J]. iMetaOmics, 2024, 1(2): e25. |
| 27 | Beier S, Thiel T, Münch T, et al. MISA-web: a web server for microsatellite prediction [J]. Bioinformatics, 2017, 33(16): 2583-2585. |
| 28 | 兰朝辉, 田徐芳, 师玉华, 等. 五月艾Artemisia indica叶绿体基因组结构及系统发育分析 [J]. 中国中药杂志, 2022, 47(22): 6058-6065. |
| Lan CH, Tian XF, Shi YH, et al. Chloroplast genome structure characteristics and phylogenetic analysis of Artemisia indica [J]. China J Chin Mater Med, 2022, 47(22): 6058-6065. | |
| 29 | 胡健鹏, 蒋露, 徐睿, 等. 安徽岳西野生茅苍术叶绿体全基因组测序及系统发育研究 [J]. 中国中药杂志, 2023, 48(1): 52-59. |
| Hu JP, Jiang L, Xu R, et al. Complete chloroplast genome sequencing and phylogeny of wild Atractylodes lancea from Yuexi, Anhui province [J]. China J Chin Mater Med, 2023, 48(1): 52-59. | |
| 30 | Li HE, Guo QQ, Xu L, et al. CPJSdraw: analysis and visualization of junction sites of chloroplast genomes [J]. PeerJ, 2023, 11: e15326. |
| 31 | Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability [J]. Mol Biol Evol, 2013, 30(4): 772-780. |
| 32 | Minh BQ, Schmidt HA, Chernomor O, et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era [J]. Mol Biol Evol, 2020, 37(5): 1530-1534. |
| 33 | Ronquist F, Teslenko M, van der Mark P, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space [J]. Syst Biol, 2012, 61(3): 539-542. |
| 34 | Plunkett GM, Downie SR. Expansion and contraction of the chloroplast inverted repeat in Apiaceae subfamily apioideae [J]. Syst Bot, 2000, 25(4): 648. |
| 35 | Zhu AD, Guo WH, Gupta S, et al. Evolutionary dynamics of the plastid inverted repeat: the effects of expansion, contraction, and loss on substitution rates [J]. New Phytol, 2016, 209(4): 1747-1756. |
| 36 | 国家药典委员会. 中华人民共和国药典-一部: 2020年版 [M]. 北京: 中国医药科技出版社, 2020: 103. |
| Pharmacopoeia Commission of the People's Republic of China. Pharmacopoeia of the People's Republic of China - Volume I: 2020 Edition [M]. Beijing: China Medical Science Press, 2020: 103. | |
| 37 | 鲁兆莉, 覃海宁, 金效华, 等. 《国家重点保护野生植物名录》调整的必要性、原则和程序 [J]. 生物多样性, 2021, 29(12): 1577-1582. |
| Lu ZL, Qin HN, Jin XH, et al. On the necessity, principle, and process of updating the List of National Key Protected Wild Plants [J]. Biodivers Sci, 2021, 29(12): 1577-1582. | |
| 38 | Guo YY, Yang JX, Bai MZ, et al. The chloroplast genome evolution of Venus slipper (Paphiopedilum): IR expansion, SSC contraction, and highly rearranged SSC regions [J]. BMC Plant Biol, 2021, 21(1): 248. |
| 39 | Wei N, Pérez-Escobar OA, Musili PM, et al. Plastome evolution in the hyperdiverse genus Euphorbia (Euphorbiaceae) using phylogenomic and comparative analyses: large-scale expansion and contraction of the inverted repeat region [J]. Front Plant Sci, 2021, 12: 712064. |
| 40 | 王爱兰, 王贵琳, 李维卫. 濒危物种珊瑚菜遗传多样性的ISSR分析 [J]. 西北植物学报, 2015, 35(8): 1541-1546. |
| Wang AL, Wang GL, Li WW. Genetic diversity of Glehnia littoralis populations revealed by ISSR molecular markers [J]. Acta Bot Boreali Occidentalia Sin, 2015, 35(8): 1541-1546. |
| [1] | LI Bin, SU Xiang-ping, LIU Chang, WANG Yu-bing, ZHANG Yong-hong, ZHOU Chao, XU Qing. Chloroplast Genome Characteristics and Phylogenetic Analysis of Scrophulariaceae [J]. Biotechnology Bulletin, 2025, 41(3): 240-254. |
| [2] | YANG Yu-qing, TAN Juan, WANG Fang, PENG Shun-li, CHEN Jie, TAN Ming-yan, LYU Mei-yan, ZHOU Fu-yu, LIU Sheng-chuan. Research and Application Progress in Chloroplast Genome of Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2024, 40(2): 20-30. |
| [3] | YIN Ming-hua, YU Huan-yuan, XIAO Xin-yi, WANG Yu-ting. Chloroplast Genomic Characterization and Phylogenetic Analysis of Colocasia esculenta L. Schoot var. cormosus cv. ‘Hongyayu’ from Jiangxi Yanshan [J]. Biotechnology Bulletin, 2023, 39(6): 233-247. |
| [4] | LIU Xiong-wei, LIU Chang, ZENG Xian-fa, YANG Xiao-ying, FENG Ting-ting, ZHAO Jie-hong, ZHOU Ying. Comparative and Phylogenetic Analyses of Complete Chloroplast Genomes in Ardisia crenata [J]. Biotechnology Bulletin, 2023, 39(1): 232-242. |
| [5] | QIAN Fang, GAO Zuo-min, HU Li-juan, WANG Hong-cheng. Characteristics of Crambe abyssinica Chloroplast Genome and Its Phylogenetic Relationship in Brassicaceae [J]. Biotechnology Bulletin, 2022, 38(6): 174-186. |
| [6] | ZHU Bin, GAN Chen-chen, WANG Hong-cheng. Characteristics of the Complete Chloroplast Genome of Dendrobium thyrsiflorum and Its Phylogenetic Relationship Analysis [J]. Biotechnology Bulletin, 2021, 37(5): 38-47. |
| [7] | LI Yu-hua, REN Yong-kang, ZHAO Xing-hua, LIU Jiang, HAN bin, WANG Chang-biao, TANG Zhao-hui. Research Progress on Chloroplast Genome of Major Gramineous Crops [J]. Biotechnology Bulletin, 2020, 36(11): 112-121. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||