








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (6): 218-228.doi: 10.13560/j.cnki.biotech.bull.1985.2024-1182
CHENG Shan1( ), WANG Hui-wei1, CHEN Chen1, ZHU Ya-jing1, LI Chun-xin1, BIE Hai2, WANG Shu-feng1, CHEN Xian-gong1, ZHANG Xiang-ge1(
), WANG Hui-wei1, CHEN Chen1, ZHU Ya-jing1, LI Chun-xin1, BIE Hai2, WANG Shu-feng1, CHEN Xian-gong1, ZHANG Xiang-ge1( )
)
Received:2024-12-05
Online:2025-06-26
Published:2025-06-30
Contact:
ZHANG Xiang-ge
E-mail:18790610677@163.com;maizezxg@163.com
CHENG Shan, WANG Hui-wei, CHEN Chen, ZHU Ya-jing, LI Chun-xin, BIE Hai, WANG Shu-feng, CHEN Xian-gong, ZHANG Xiang-ge. Cloning of MYB Transcription Factor Gene CeMYB154 and Analysis of Salt Tolerance Function in Cyperus esculentus[J]. Biotechnology Bulletin, 2025, 41(6): 218-228.
| 引物名称 Primer name | 引物序列 Primer sequence (5′‒3′) | 用途 Purpose |
|---|---|---|
| MYB154-F | ATGGAGGAAGTGGCATGGAG | 基因克隆 Gene clone |
| MYB154-R | CTAGTATAAAGCAAATTCTGCTGCTTGC | |
| 2300-F | gtacccggggatcctctagagtcgacATGGAGGAAGTGGCATGGAG | 转基因植株检测 Transgenic plant detection |
| 2300-R | ttgctcaccatggtactagtgtcgacGTATAAAGCAAATTCTGCTGCTTGC | |
| GFP-F | ctatttacaattacggatccATGGAGGAAGTGGCATGGAG | 亚细胞定位 Subcellular localization |
| GFP-R | agatcctcctccggagaccTAAAGCAAATTCTGCTGC | |
| BD-F | atggccatggaggccgaattcATGGAGGAAGTGGCATGGAG | 转录激活活性检测 Transcriptional activation activity assays |
| BD-R | tcgacggatccccgggaattcGTATAAAGCAAATTCTGCTGCTTGC | |
| QP-F | GATGGAATTCTGTTGCT | 荧光定量PCR引物 Fluorescent quantitative PCR primer |
| QP-R | CTTTATCTCGTTGTCGG | |
| CetActin-F | ACTCTGGTGATGGTGTGAGC | 油莎豆内参引物 Internal primer of Cyperus esculentus |
| CetActin-R | CCCTCTCTCCGTCAGGATCT | |
| AtActin-F | GGTAACATTGTGCTCAGTGGTGG | 拟南芥内参引物 Internal primer of Arabidopsis thaliana |
| AtActin-R | AACGACCTTAATCTTCATGCTGC |
Table 1 Primer sequences used in this study
| 引物名称 Primer name | 引物序列 Primer sequence (5′‒3′) | 用途 Purpose |
|---|---|---|
| MYB154-F | ATGGAGGAAGTGGCATGGAG | 基因克隆 Gene clone |
| MYB154-R | CTAGTATAAAGCAAATTCTGCTGCTTGC | |
| 2300-F | gtacccggggatcctctagagtcgacATGGAGGAAGTGGCATGGAG | 转基因植株检测 Transgenic plant detection |
| 2300-R | ttgctcaccatggtactagtgtcgacGTATAAAGCAAATTCTGCTGCTTGC | |
| GFP-F | ctatttacaattacggatccATGGAGGAAGTGGCATGGAG | 亚细胞定位 Subcellular localization |
| GFP-R | agatcctcctccggagaccTAAAGCAAATTCTGCTGC | |
| BD-F | atggccatggaggccgaattcATGGAGGAAGTGGCATGGAG | 转录激活活性检测 Transcriptional activation activity assays |
| BD-R | tcgacggatccccgggaattcGTATAAAGCAAATTCTGCTGCTTGC | |
| QP-F | GATGGAATTCTGTTGCT | 荧光定量PCR引物 Fluorescent quantitative PCR primer |
| QP-R | CTTTATCTCGTTGTCGG | |
| CetActin-F | ACTCTGGTGATGGTGTGAGC | 油莎豆内参引物 Internal primer of Cyperus esculentus |
| CetActin-R | CCCTCTCTCCGTCAGGATCT | |
| AtActin-F | GGTAACATTGTGCTCAGTGGTGG | 拟南芥内参引物 Internal primer of Arabidopsis thaliana |
| AtActin-R | AACGACCTTAATCTTCATGCTGC |

Fig. 1 Cloning and bioinformatics analysis of CeMYB154A: CeMYB154 gene cloning. B: The expressions of CeMYB154 in Cyperus esculentus roots under different concentrations of NaCl treatment. C: CeMYB154 protein domain analysis. D: CeMYB154 subcellular localization prediction. E: Multi-species sequence alignment. Ce: Cyperus esculentus. Ah:: Arachis hypogaea. Gm: Glycine max. Zm: Zea mays. Bna: Brassica napus. At: Arabidopsis thaliana. Os: Oryza sativa. St: Solanum tuberosum. Ib: Ipomoea batatas. ** indicates a significant difference at the P<0.01 level with WT as control, the same below
| 顺式作用元件 Cis-acting element | 序列 Sequence (5′‒3′) | 功能 Function | 数量 Quantity |
|---|---|---|---|
| CAAT-box | CAAT | 启动子和增强子区域共同的顺式作用元件 | 23 |
| TATA-box | TATATA; ATATAT | 转录起始-30附近的核心启动子元件 | 156 |
| MYB-like | TAACCA | MYB识别和结合元件 | 1 |
| MRE | AACCTAA | MYB响应元件 | 1 |
| TCCC-motif | TCTCCCT | 光响应元件 | 1 |
| GT1-motif | GGTTAAT | 光响应元件 | 5 |
| G-box | CACGTC | 光响应元件 | 1 |
| GATA-motif | GATAGGG | 光响应元件 | 1 |
| AAGAA-motif | GAAAGAA | 脱落酸响应元件 | 2 |
| ABRE | ACGTG | 脱落酸响应元件 | 1 |
| TGA-box | TGACGTAA | 生长素响应元件 | 1 |
| TGACG-motif | TGACG | 茉莉酸甲酯响应元件 | 2 |
| ARE | AAACCA | 厌氧诱导调节元件 | 3 |
Table 2 Analysis of cis-acting elements in CeMYB154 promoter
| 顺式作用元件 Cis-acting element | 序列 Sequence (5′‒3′) | 功能 Function | 数量 Quantity |
|---|---|---|---|
| CAAT-box | CAAT | 启动子和增强子区域共同的顺式作用元件 | 23 |
| TATA-box | TATATA; ATATAT | 转录起始-30附近的核心启动子元件 | 156 |
| MYB-like | TAACCA | MYB识别和结合元件 | 1 |
| MRE | AACCTAA | MYB响应元件 | 1 |
| TCCC-motif | TCTCCCT | 光响应元件 | 1 |
| GT1-motif | GGTTAAT | 光响应元件 | 5 |
| G-box | CACGTC | 光响应元件 | 1 |
| GATA-motif | GATAGGG | 光响应元件 | 1 |
| AAGAA-motif | GAAAGAA | 脱落酸响应元件 | 2 |
| ABRE | ACGTG | 脱落酸响应元件 | 1 |
| TGA-box | TGACGTAA | 生长素响应元件 | 1 |
| TGACG-motif | TGACG | 茉莉酸甲酯响应元件 | 2 |
| ARE | AAACCA | 厌氧诱导调节元件 | 3 |

Fig. 2 Subcellular localizationEGFP indicates green fluorescence; Cy5 indicates red fluorescence; TD indicates bright field; ALL indicates the superposition of three light sources
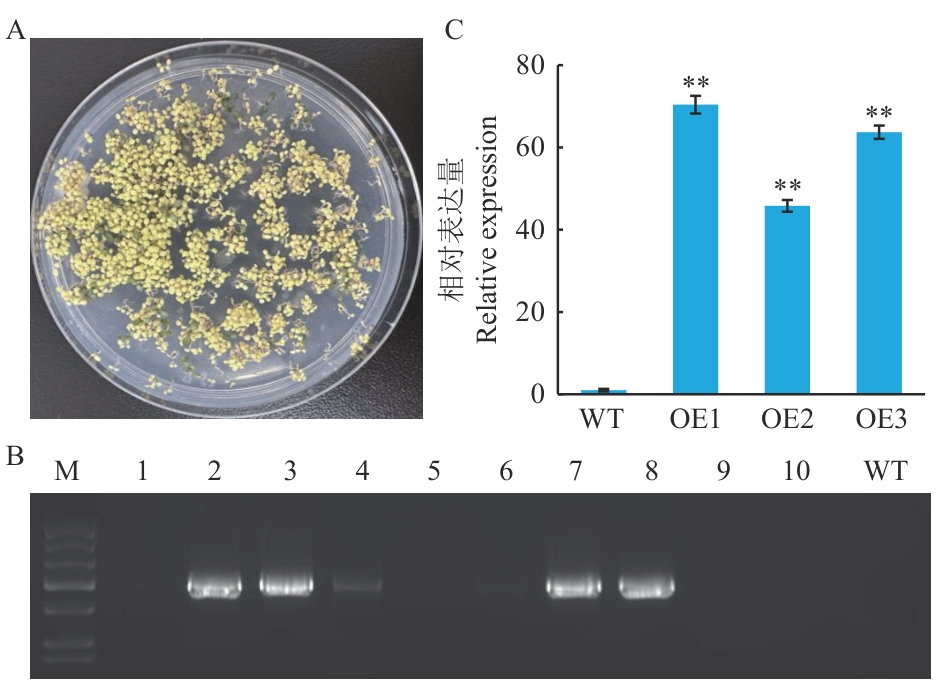
Fig. 4 Identification of transgenic A. thaliana plantsA: Hygromycin screening T1 transgenic vaccines. B: PCR identification of T1 transgenic plants (M is DL2000 marker, 1‒10 are transgenic plants, and WT is wild-type plants). C: Detection of CeMYB154 expression in T2 transgenic A. thaliana plants, OE1, OE2 and OE3 indicate an independent T2 generation transgenic plant, respectively, the same below

Fig. 5 Phenotype of salt-tolerant in CeMYB154 transgenic A. thaliana plantsA: Effect of salt stress on the seedling growth of A. thaliana plants. B: Effect of salt stress on the root growth of transgenic A. thaliana plants. C: Root length of WT and transgenic A.thaliana plants under salt stress
| 1 | 张玉娟, 黎冬华, 宫慧慧, 等. 芝麻NAC转录因子基因SiNAC77的克隆及耐盐功能分析 [J]. 生物技术通报, 2023, 39(11): 308-317. |
| Zhang YJ, Li DH, Gong HH, et al. Cloning and salt-tolerance analysis of NAC transcription factor SiNAC77 from Sesamum indicum L [J]. Biotechnol Bull, 2023, 39(11): 308-317. | |
| 2 | 杨帆, 朱文学. 油莎豆研究现状及展望 [J]. 粮食与油脂, 2020, 33(7): 4-6. |
| Yang F, Zhu WX. Research status and prospect of Cyperus esculentus [J]. Cereals Oils, 2020, 33(7): 4-6. | |
| 3 | 刘玉兰, 王小宁, 舒垚, 等. 不同产地油莎豆性状及组成分析研究 [J]. 中国油脂, 2020, 45(8): 125-129. |
| Liu YL, Wang XN, Shu Y, et al. Character and composition of Cyperus esculentus from different origins [J]. China Oils Fats, 2020, 45(8): 125-129. | |
| 4 | 王会伟, 张向歌, 李春鑫, 等. 油莎豆耐盐性评估及盐胁迫下幼苗根系转录组学分析 [J]. 作物学报, 2023, 49(7): 1882-1894. |
| Wang HW, Zhang XG, Li CX, et al. Evaluation of salt tolerance in Cyperus esculentus and transcriptomic analysis of seedling roots under salt stress [J]. Acta Agron Sin, 2023, 49(7): 1882-1894. | |
| 5 | 周扬, 王鹏, 李雨欣. 植物耐盐分子机制研究进展 [J]. 广东农业科学, 2023, 50(10): 97-109. |
| Zhou Y, Wang P, Li YX. Research progress on molecular mechanism of plant salt tolerance [J]. Guangdong Agric Sci, 2023, 50(10): 97-109. | |
| 6 | 冯凯月, 赵鑫焱, 李子妍, 等. 植物响应盐碱胁迫的分子机制研究进展 [J]. 生物技术通报, 2024, 40(10): 122-138. |
| Feng KY, Zhao XY, Li ZY, et al. Research advances in the molecular mechanisms of plant response to saline-alkali stress [J]. Biotechnol Bull, 2024, 40(10): 122-138. | |
| 23 | 李星伟, 王庆江, 吴宇童, 等. 苹果MdMYB113基因响应高盐胁迫的功能鉴定 [J]. 植物生理学报, 2021, 57(3): 579-588. |
| Li XW, Wang QJ, Wu YT, et al. Functional identification of MdMYB113 gene in apple in response to high salt stress [J]. Plant Physiol J, 2021, 57(3): 579-588. | |
| 24 | Cao YP, Han YH, Li DH, et al. MYB transcription factors in Chinese pear (Pyrus bretschneideri rehd.): genome-wide identification, classification, and expression profiling during fruit development [J]. Front Plant Sci, 2016, 7: 577. |
| 25 | Zhang ZJ, Zhang L, Liu Y, et al. Identification and expression analysis of R2R3-MYB family genes associated with salt tolerance in Cyclocarya paliurus [J]. Int J Mol Sci, 2022, 23(7): 3429. |
| 26 | 张红梅, 刘晓庆, 陈华涛, 等. 大豆转录因子GmWRKY58亚细胞定位及在非生物胁迫下的表达分析 [J]. 华北农学报, 2018, 33(2): 49-57. |
| Zhang HM, Liu XQ, Chen HT, et al. Subcellular localization and expression analysis of a soybean transcription factor GmWRKY58 in response to abiotic stresses [J]. Acta Agric Boreali Sin, 2018, 33(2): 49-57. | |
| 27 | 陈彧, 邢文婷, 李雨欣, 等. 铁皮石斛DcNAC1基因克隆、表达及转录自激活活性分析 [J]. 南方农业学报, 2023, 54(6): 1612-1621. |
| Chen Y, Xing WT, Li YX, et al. Cloning, expression and trans-activation activity analysis of DcNAC1 gene from Dendrobium catenatum [J]. J South Agric, 2023, 54(6): 1612-1621. | |
| 28 | Dossa K, Mmadi MA, Zhou R, et al. Ectopic expression of the sesame MYB transcription factor SiMYB305 promotes root growth and modulates ABA-mediated tolerance to drought and salt stresses in Arabidopsis [J]. AoB Plants, 2019, 12(1): plz081. |
| 29 | 胡雅丹, 伍国强, 刘晨, 等. MYB转录因子在调控植物响应逆境胁迫中的作用 [J]. 生物技术通报, 2024, 40(6): 5-22. |
| Hu YD, Wu GQ, Liu C, et al. Roles of MYB transcription factor in regulating the responses of plants to stress [J]. Biotechnol Bull, 2024, 40(6): 5-22. | |
| 30 | He YX, Dong YS, Yang XD, et al. Functional activation of a novel R2R3-MYB protein gene, GmMYB68, confers salt-alkali resistance in soybean (Glycine max L.) [J]. Genome, 2020, 63(1): 13-26. |
| 7 | Zhang S, Wu QR, Liu LL, et al. Osmotic stress alters circadian cytosolic Ca2+ oscillations and OSCA1 is required in circadian gated stress adaptation [J]. Plant Signal Behav, 2020, 15(12): 1836883. |
| 8 | Jiang ZH, Zhou XP, Tao M, et al. Plant cell-surface GIPC sphingolipids sense salt to trigger Ca2+ influx [J]. Nature, 2019, 572(7769): 341-346. |
| 9 | Chen K, Gao JH, Sun SJ, et al. BONZAI proteins control global osmotic stress responses in plants [J]. Curr Biol, 2020, 30(24): 4815-4825.e4. |
| 10 | Yan JW, Liu Y, Yan JW, et al. The salt-activated CBF1/CBF2/CBF3-GALS1 module fine-tunes galactan-induced salt hypersensitivity in Arabidopsis [J]. J Integr Plant Biol, 2023, 65(8): 1904-1917. |
| 11 | Gao F, Zhou J, Deng RY, et al. Overexpression of a Tartary buckwheat R2R3-MYB transcription factor gene, FtMYB9, enhances tolerance to drought and salt stresses in transgenic Arabidopsis [J]. J Plant Physiol, 2017, 214: 81-90. |
| 12 | Le Hir R, Castelain M, Chakraborti D, et al. AtbHLH68 transcription factor contributes to the regulation of ABA homeostasis and drought stress tolerance in Arabidopsis thaliana [J]. Physiol Plant, 2017, 160(3): 312-327. |
| 13 | Raineri J, Wang SH, Peleg Z, et al. The rice transcription factor OsWRKY47 is a positive regulator of the response to water deficit stress [J]. Plant Mol Biol, 2015, 88(4/5): 401-413. |
| 14 | Dubos C, Stracke R, Grotewold E, et al. MYB transcription factors in Arabidopsis [J]. Trends Plant Sci, 2010, 15(10): 573-581. |
| 15 | Wang YJ, Zhang Y, Fan CJ, et al. Genome-wide analysis of MYB transcription factors and their responses to salt stress in Casuarina equisetifolia [J]. BMC Plant Biol, 2021, 21(1): 328. |
| 16 | Liao Y, Zou HF, Wang HW, et al. Soybean GmMYB76, GmMYB92, and GmMYB177 genes confer stress tolerance in transgenic Arabidopsis plants [J]. Cell Res, 2008, 18(10): 1047-1060. |
| 17 | Yoo JH, Park CY, Kim JC, et al. Direct interaction of a divergent CaM isoform and the transcription factor, MYB2, enhances salt tolerance in Arabidopsis [J]. J Biol Chem, 2005, 280(5): 3697-3706. |
| 18 | Zhu N, Cheng SF, Liu XY, et al. The R2R3-type MYB gene OsMYB91 has a function in coordinating plant growth and salt stress tolerance in rice [J]. Plant Sci, 2015, 236: 146-156. |
| 19 | Zhang X, Chen LC, Shi QH, et al. SlMYB102, an R2R3-type MYB gene, confers salt tolerance in transgenic tomato [J]. Plant Sci, 2020, 291: 110356. |
| 20 | Zhao YY, Yang ZE, Ding YP, et al. Over-expression of an R2R3 MYB gene, GhMYB73, increases tolerance to salt stress in transgenic Arabidopsis [J]. Plant Sci, 2019, 286: 28-36. |
| 21 | Dong W, Liu XJ, Li DL, et al. Transcriptional profiling reveals that a MYB transcription factor MsMYB4 contributes to the salinity stress response of alfalfa [J]. PLoS One, 2018, 13(9): e0204033. |
| 22 | Du BY, Liu H, Dong KT, et al. Over-expression of an R2R3 MYB gene, MdMYB108L, enhances tolerance to salt stress in transgenic plants [J]. Int J Mol Sci, 2022, 23(16): 9428. |
| 31 | Zhang WX, Wang N, Yang JT, et al. The salt-induced transcription factor GmMYB84 confers salinity tolerance in soybean [J]. Plant Sci, 2020, 291: 110326. |
| 32 | Du YT, Zhao MJ, Wang CT, et al. Identification and characterization of GmMYB118 responses to drought and salt stress [J]. BMC Plant Biol, 2018, 18(1): 320. |
| 33 | Pi EX, Xu J, Li HH, et al. Enhanced salt tolerance of rhizobia-inoculated soybean correlates with decreased phosphorylation of the transcription factor GmMYB183 and altered flavonoid biosynthesis [J]. Mol Cell Proteomics, 2019, 18(11): 2225-2243. |
| 34 | Pi EX, Zhu CM, Fan W, et al. Quantitative phosphoproteomic and metabolomic analyses reveal GmMYB173 optimizes flavonoid metabolism in soybean under salt stress [J]. Mol Cell Proteomics, 2018, 17(6): 1209-1224. |
| 35 | Wang C, Wang LJ, Lei J, et al. IbMYB308, a sweet potato R2R3-MYB gene, improves salt stress tolerance in transgenic tobacco [J]. Genes, 2022, 13(8): 1476. |
| 36 | Hossain MA, Bhattacharjee S, Armin SM, et al. Hydrogen peroxide priming modulates abiotic oxidative stress tolerance: insights from ROS detoxification and scavenging [J]. Front Plant Sci, 2015, 6: 420. |
| 37 | Liu P, Wu XL, Gong BB, et al. Review of the mechanisms by which transcription factors and exogenous substances regulate ROS metabolism under abiotic stress [J]. Antioxidants, 2022, 11(11): 2106. |
| 38 | Li WH, Zhong JL, Zhang LH, et al. Overexpression of a Fragaria vesca MYB transcription factor gene (FvMYB82) increases salt and cold tolerance in Arabidopsis thaliana [J]. Int J Mol Sci, 2022, 23(18): 10538. |
| 39 | 程钰鑫. R2R3 MYB转录因子MYB71、MYB79和MYB121对拟南芥ABA及干旱胁迫响应的调控 [D]. 长春: 东北师范大学, 2022. |
| Cheng YX. Regulation of R2R3 MYB transcription factors MYB71, MYB79 and MYB121 on ABA and drought stress response in Arabidopsis thaliana [D]. Changchun: Northeast Normal University, 2022. |
| [1] | XU Hui-zhen, SHANTWANA Ghimire, RAJU Kharel, YUE Yun, SI Huai-jun, TANG Xun. Analysis of the Potato SUMO E3 Ligase Gene Family and Cloning and Expression Pattern of StSIZ1 [J]. Biotechnology Bulletin, 2025, 41(6): 119-129. |
| [2] | WU Ya, YAO Run, YANG Han-ting, LIU Wei, YANG Shuai, SONG Chi, CHEN Shi-lin. Genome-wide Identification and Expression Analysis of SDR Gene Family in Mentha suaveolens ‘Variegata’ [J]. Biotechnology Bulletin, 2025, 41(5): 175-185. |
| [3] | LI Zhi-qiang, LUO Zheng-qian, XU Lin-li, ZHOU Guo-hui, QU Si-yu, LIU En-liang, XU Dong-ting. Identification of the Soybean R2R3-MYB Gene Family Based on the T2T Genome and Their Expression Analysis under Drought and Salt Stress [J]. Biotechnology Bulletin, 2025, 41(5): 141-152. |
| [4] | YANG Chun, WANG Xiao-qian, WANG Hong-jun, CHAO Yue-hui. Cloning, Subcellular Localization and Expression Analysis of MtZHD4 Gene from Medicago truncatula [J]. Biotechnology Bulletin, 2025, 41(5): 244-254. |
| [5] | SONG Hui-yang, SU Bao-jie, LI Jing-hao, MEI Chao, SONG Qian-na, CUI Fu-zhu, FENG Rui-yun. Cloning and Functional Analysis of the StAS2-15 Gene in Potato under Salt Stress [J]. Biotechnology Bulletin, 2025, 41(5): 119-128. |
| [6] | BAN Qiu-yan, ZHAO Xin-yue, CHI Wen-jing, LI Jun-sheng, WANG Qiong, XIA Yao, LIANG Li-yun, HE Wei, LI Ye-yun, ZHAO Guang-shan. Cloning of Phytochrome Interaction Factor CsPIF3a and Its Response to Light and Temperature Stress in Camellia sinensis [J]. Biotechnology Bulletin, 2025, 41(4): 256-265. |
| [7] | LU Yong-jie, XIA Hai-qian, LI Yong-ling, ZHANG Wen-jian, YU Jing, ZHAO Hui-na, WANG Bing, XU Ben-bo, LEI Bo. Cloning and Expression Analysis of AP2/ERF Transcription Factor NtESR2 in Nicotiana tabacum [J]. Biotechnology Bulletin, 2025, 41(4): 266-277. |
| [8] | LIU Tao, WANG Zhi-qi, WU Wen-bo, SHI Wen-ting, WANG Chao-nan, DU Chong, YANG Zhong-min. Identification and Expression Analysis of the GRAM Gene Family in Potato [J]. Biotechnology Bulletin, 2025, 41(4): 145-155. |
| [9] | LI Cai-xia, LI Yi, MU Hong-xiu, LIN Jun-xuan, BAI Long-qiang, SUN Mei-hua, MIAO Yan-xiu. Identification and Bioinformatics Analysis of the bHLH Transcription Factor Family in Cucurbita moschata Duch. [J]. Biotechnology Bulletin, 2025, 41(1): 186-197. |
| [10] | YUAN Liu-jiao, HUANG Wen-lin, CHEN Chong-zhi, LIANG Min, HUANG Zi-qi, CHEN Xue-xue, CHEN Ri-Meng, WANG Li-yun. Effects of Salt Stress on Physiological Characteristics, Ultrastructure and Medicinal Components of Pogostemon cablin Leaves [J]. Biotechnology Bulletin, 2025, 41(1): 230-239. |
| [11] | QIAO Yan, YANG Fang, REN Pan-rong, QI Wei-liang, AN Pei-pei, LI Qian, LI Dan, XIAO Jun-fei. Cloning and Function Analysis of the ScDHNS Gene of Crotonase/Enoyl-CoA Superfamily from a Wild Potato Species [J]. Biotechnology Bulletin, 2024, 40(9): 92-103. |
| [12] | XING Li-nan, ZHANG Yan-fang, GE Ming-ran, ZHAO Ling-min, CHEN Yan, HUO Xiu-wen. Analysis of DoWRKY40 Gene Expression Characteristics and Screening of Interacting Proteins in Yam [J]. Biotechnology Bulletin, 2024, 40(8): 118-128. |
| [13] | WU Shuai, XIN Yan-ni, MAI Chun-hai, MU Xiao-ya, WANG Min, YUE Ai-qin, ZHAO Jin-zhong, WU Shen-jie, DU Wei-jun, WANG Li-xiang. Genome-wide Identification and Stress Response Analysis of Soybean GS Gene Family [J]. Biotechnology Bulletin, 2024, 40(8): 63-73. |
| [14] | LIN Tong, YUAN Cheng, DONG Chen-wen-hua, ZENG Meng-qiong, YANG Yan, MAO Zi-chao, LIN Chun. Screening and Functional Analysis of Gene CqSTK Associated with Gametophyte Development of Quinoa [J]. Biotechnology Bulletin, 2024, 40(8): 83-94. |
| [15] | HU Ya-dan, WU Guo-qiang, LIU Chen, WEI Ming. Roles of MYB Transcription Factor in Regulating the Responses of Plants to Stress [J]. Biotechnology Bulletin, 2024, 40(6): 5-22. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||