








Biotechnology Bulletin ›› 2026, Vol. 42 ›› Issue (1): 139-149.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0557
Previous Articles Next Articles
NI Ying1( ), LI Lei1, WANG Jin-xuan1, MA Bo1, MENG Xin2, LENG Ping-sheng1,3, WU Jing1,3(
), LI Lei1, WANG Jin-xuan1, MA Bo1, MENG Xin2, LENG Ping-sheng1,3, WU Jing1,3( ), HU Zeng-hui1,3(
), HU Zeng-hui1,3( )
)
Received:2025-05-31
Online:2026-01-26
Published:2026-02-04
Contact:
WU Jing, HU Zeng-hui
E-mail:18810602920@163.com;wjmxy1988@126.com;buahuzenghui@163.com
NI Ying, LI Lei, WANG Jin-xuan, MA Bo, MENG Xin, LENG Ping-sheng, WU Jing, HU Zeng-hui. Cloning and Functional Analysis of So4CL Gene in Syringa oblata[J]. Biotechnology Bulletin, 2026, 42(1): 139-149.
引物用途 Primer function | 引物名称 Primer name | 引物序列 Primer sequence (5′‒3′) |
|---|---|---|
克隆基因 Gene cloning | So4CL-F So4CL-R | ATGGAGACTAAGACGACGCAAGAAGA TCAATTGGGAACACCTGCAGCTAATC |
实时荧光定量PCR RT-qPCR | qSoActin-F qSoActin-R qSo4CL-F qSo4CL-R qSoPAL-F qSoPAL-R qSoCHS-F qSoCHS-R qSoDFR-F qSoDFR-R qSoUFGT-F qSoUFGT-R | TGGAATGTGCTGAGAGATGC TGCTGACCGTATGAGCAAAG CTTGCGGTCTGTCTTGTCTG TTCCGTCAAACCATAGCCCT TGGACTATGGCTTCAAGGGGR CTCGCCGAAATCAAACCCAA TCTCGTAGTGTGCTCGGAAA GTCAGAACCCACAATCACGG GCATTGGAAGCGGCTAAAGA CCAGTGATAGGCGAAAGTGC TACCACCAGAATAGAGT AGAAGTATTGCCAGAAG |
基因过表达 Gene overexpression | PRI101-So4CL-F PRI101-So4CL-R | ATGCCCGTCGACCCCGGGGGCGGAACTCAACCAGC CTCACCATGGATCCGGTACCTAGTTTAGATACGGCAAGCTTTATTAGATCCTTCC |
基因沉默 Gene silencing | TRV2-So4CL-F TRV2-So4CL-R | GTCCAGTCCTGGCCTCGTCGGCCATGGCGGAACTCAACCAGC GACCACAAGTGGCCAGACTGGCCAAGCTGAGCTGATGCCCG |
So4CL-SubN-F So4CL-SubN-R | AGTGGTCTCTGTCCAGTCCTATGGAGACTAAGACGACGCAAGAAGA GGTCTCAGCAGACCACAAGTATTGGGAACACCTGCAGCTAATC |
Table 1 Primer sequences
引物用途 Primer function | 引物名称 Primer name | 引物序列 Primer sequence (5′‒3′) |
|---|---|---|
克隆基因 Gene cloning | So4CL-F So4CL-R | ATGGAGACTAAGACGACGCAAGAAGA TCAATTGGGAACACCTGCAGCTAATC |
实时荧光定量PCR RT-qPCR | qSoActin-F qSoActin-R qSo4CL-F qSo4CL-R qSoPAL-F qSoPAL-R qSoCHS-F qSoCHS-R qSoDFR-F qSoDFR-R qSoUFGT-F qSoUFGT-R | TGGAATGTGCTGAGAGATGC TGCTGACCGTATGAGCAAAG CTTGCGGTCTGTCTTGTCTG TTCCGTCAAACCATAGCCCT TGGACTATGGCTTCAAGGGGR CTCGCCGAAATCAAACCCAA TCTCGTAGTGTGCTCGGAAA GTCAGAACCCACAATCACGG GCATTGGAAGCGGCTAAAGA CCAGTGATAGGCGAAAGTGC TACCACCAGAATAGAGT AGAAGTATTGCCAGAAG |
基因过表达 Gene overexpression | PRI101-So4CL-F PRI101-So4CL-R | ATGCCCGTCGACCCCGGGGGCGGAACTCAACCAGC CTCACCATGGATCCGGTACCTAGTTTAGATACGGCAAGCTTTATTAGATCCTTCC |
基因沉默 Gene silencing | TRV2-So4CL-F TRV2-So4CL-R | GTCCAGTCCTGGCCTCGTCGGCCATGGCGGAACTCAACCAGC GACCACAAGTGGCCAGACTGGCCAAGCTGAGCTGATGCCCG |
So4CL-SubN-F So4CL-SubN-R | AGTGGTCTCTGTCCAGTCCTATGGAGACTAAGACGACGCAAGAAGA GGTCTCAGCAGACCACAAGTATTGGGAACACCTGCAGCTAATC |
蛋白名称 Protein name | 分子式 Formula | 相对分子质量 Molecular weight (Da) | 总原子数 Total number of atoms | 酸碱性氨基酸 Acid-base amino acid | 理论等电点 Theoretical pI | 脂溶指数 Aliphatic index | |
|---|---|---|---|---|---|---|---|
| Asp + Glu | Arg + Lys | ||||||
| So4CL | C2716H4383N709O804S18 | 60 411.03 | 8 630 | 57 | 61 | 8.54 | 102.28 |
Table 2 Analysis of physical and chemical properties of So4CL protein
蛋白名称 Protein name | 分子式 Formula | 相对分子质量 Molecular weight (Da) | 总原子数 Total number of atoms | 酸碱性氨基酸 Acid-base amino acid | 理论等电点 Theoretical pI | 脂溶指数 Aliphatic index | |
|---|---|---|---|---|---|---|---|
| Asp + Glu | Arg + Lys | ||||||
| So4CL | C2716H4383N709O804S18 | 60 411.03 | 8 630 | 57 | 61 | 8.54 | 102.28 |

Fig. 3 Alignment and analysis of So4CL with 4CL protein sequences of other speciesThe black, red, and blue parts indicate homology =100%, ≥75%, ≥50% respectively. Red boxes are SSGTTGLPKGV conserved motifs, and green boxes are GEXXIXG conserved motifs
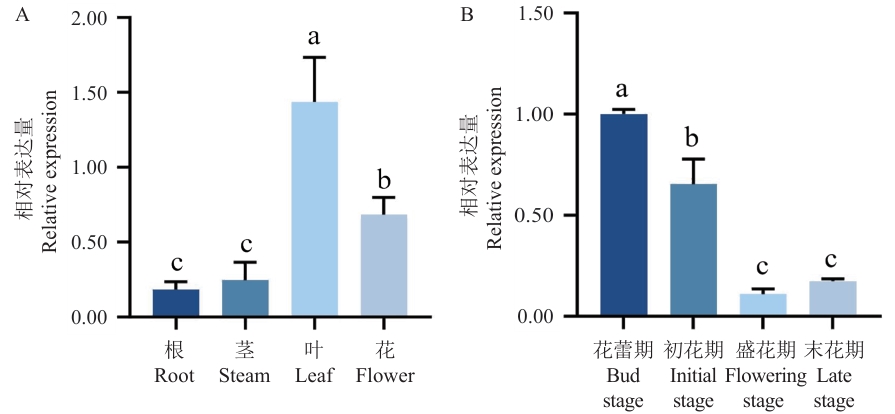
Fig. 5 Expression patterns of So4CL at different organs (A) and in different flower developmental stages (B)Different lowercase letters indicate significant differences at the P<0.05 level
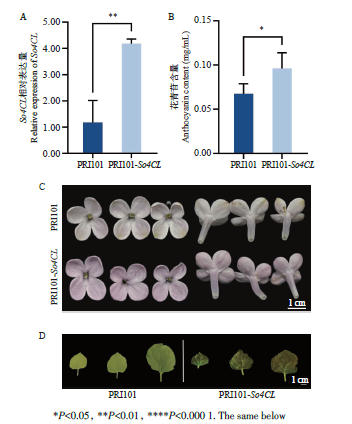
Fig. 7 Expressions of So4CL (A), anthocyanin contents (B), and petal color (C) in So4CL-overexpressed S. oblata and leaf color in So4CL-overexpressing N. benthamian (D)
| [1] | Ma B, Wu J, Shi TL, et al. Lilac (Syringa oblata) genome provides insights into its evolution and molecular mechanism of petal color change [J]. Commun Biol, 2022, 5(1): 686. |
| [2] | 宋毓晔, 王渝, 朱千林, 等. 植物中花色苷来源及提取方法研究进展 [J]. 食品研究与开发, 2022, 43(16): 199-208. |
| Song YY, Wang Y, Zhu QL, et al. Research progress on sources and extraction methods of plant-based anthocyanin [J]. Food Res Dev, 2022, 43(16): 199-208. | |
| [3] | Zhu J, Wang YZ, Wang QY, et al. The combination of DNA methylation and positive regulation of anthocyanin biosynthesis by MYB and bHLH transcription factors contributes to the petal blotch formation in Xibei tree peony [J]. Hortic Res, 2023, 10(6): uhad100. |
| [4] | Deng CY, Wang JY, Lu CF, et al. CcMYB6-1 and CcbHLH1, two novel transcription factors synergistically involved in regulating anthocyanin biosynthesis in cornflower [J]. Plant Physiol Biochem, 2020, 151: 271-283. |
| [5] | 黄玲, 胡先梅, 梁泽慧, 等. 郁金香花青素合成酶基因TgANS的克隆与功能鉴定 [J]. 园艺学报, 2022, 49(9): 1935-1944. |
| Huang L, Hu XM, Liang ZH, et al. Cloning and function identification of anthocyanidin synthase gene TgANS in Tulipa gesneriana [J]. Acta Hortic Sin, 2022, 49(9): 1935-1944. | |
| [6] | 刘玥, 李月庆, 孟祥宇, 等. 大花君子兰查尔酮合酶基因CmCHS的克隆及其功能验证 [J]. 园艺学报, 2021, 48(10): 1847-1858. |
| Liu Y, Li YQ, Meng XY, et al. Cloning and functional characterization of chalcone synthase genes (CmCHS) from Clivia miniata [J]. Acta Hortic Sin, 2021, 48(10): 1847-1858. | |
| [7] | Osorio-Guarín JA, Gopaulchan D, Quanckenbush C, et al. Comparative transcriptomic analysis reveals key components controlling spathe color in Anthurium andraeanum (Hort.) [J]. PLoS One, 2021, 16(12): e0261364. |
| [8] | Rausher MD, Miller RE, Tiffin P. Patterns of evolutionary rate variation among genes of the anthocyanin biosynthetic pathway [J]. Mol Biol Evol, 1999, 16(2): 266-274. |
| [9] | Chen XH, Wang HT, Li XY, et al. Molecular cloning and functional analysis of 4-Coumarate: CoA ligase 4 (4CL-like 1) from Fraxinus mandshurica and its role in abiotic stress tolerance and cell wall synthesis [J]. BMC Plant Biol, 2019, 19(1): 231. |
| [10] | Abdullah-Zawawi MR, Ahmad-Nizammuddin NF, Govender N, et al. Comparative genome-wide analysis of WRKY, MADS-box and MYB transcription factor families in Arabidopsis and rice [J]. Sci Rep, 2021, 11(1): 19678. |
| [11] | 申晚霞, 王志彬, 薛杨, 等. 柑橘4CL基因家族的结构及其功能分析 [J]. 园艺学报, 2019, 46(6): 1068-1078. |
| Shen WX, Wang ZB, Xue Y, et al. Characterization of 4-coumarate: CoA ligase (4CL) gene family in Citrus [J]. Acta Hortic Sin, 2019, 46(6): 1068-1078. | |
| [12] | Ma JY, Zuo DJ, Zhang XD, et al. Genome-wide identification analysis of the 4-Coumarate: CoA ligase (4CL) gene family expression profiles in Juglans regia and its wild relatives J. Mandshurica resistance and salt stress [J]. BMC Plant Biol, 2024, 24(1): 211. |
| [13] | 陈心仪, 吴成英, 贺海皓, 等. 滇水金凤4CL基因的克隆及表达分析 [J]. 福建农业学报, 2024, 39(1): 40-48. |
| Chen XY, Wu CY, He HH, et al. Cloning and expression of 4CLs in Impatiens uliginosa [J]. Fujian J Agric Sci, 2024, 39(1): 40-48. | |
| [14] | Li Y, Kim JI, Pysh L, et al. Four isoforms of Arabidopsis 4-coumarate: CoA ligase have overlapping yet distinct roles in phenylpropanoid metabolism [J]. Plant Physiol, 2015, 169(4): 2409-2421. |
| [15] | 张蕾, 林晓, 罗赟, 等. RNAi沉默Fa4CL基因对草莓果实花色苷代谢的影响 [J]. 果树学报, 2015, 32(3): 434-439, 522. |
| Zhang L, Lin X, Luo Y, et al. Influences of RNAi-induced Fa4CL silencing on anthocyanin metabolism in strawberry fruit [J]. J Fruit Sci, 2015, 32(3): 434-439, 522. | |
| [16] | Zhou XH, Jacobs TB, Xue LJ, et al. Exploiting SNPs for biallelic CRISPR mutations in the outcrossing woody perennial Populus reveals 4-coumarate: CoA ligase specificity and redundancy [J]. New Phytol, 2015, 208(2): 298-301. |
| [17] | 陈锐, 马道铖, 徐睆, 等. 野蔷薇4CL基因的克隆及表达分析 [J]. 北方园艺, 2022(17): 69-78. |
| Chen R, Ma DC, Xu H, et al. Cloning, sequencing and expression analysis of Rosa multiflora Rm4CL gene [J]. North Hortic, 2022(17): 69-78. | |
| [18] | 陈蒙, 刘海峰. 山葡萄4CL基因克隆与表达分析 [J]. 扬州大学学报: 农业与生命科学版, 2019, 40(5): 20-25. |
| Chen M, Liu HF. Expression of the 4CL gene isolated from Vitis amurensis in E. coli [J]. J Yangzhou Univ Agric Life Sci Ed, 2019, 40(5): 20-25. | |
| [19] | Chen XH, Su WL, Zhang H, et al. Fraxinus mandshurica 4-coumarate-CoA ligase 2 enhances drought and osmotic stress tolerance of tobacco by increasing coniferyl alcohol content [J]. Plant Physiol Biochem, 2020, 155: 697-708. |
| [20] | 徐靖, 林延慧, 王效宁, 等. 甘薯4-香豆酸辅酶A连接酶基因的生物信息学鉴定和表达分析 [J]. 西北植物学报, 2020, 40(4): 581-587. |
| Xu J, Lin YH, Wang XN, et al. Bioinformatic identification and expression analysis of 4CL genes in Ipomoea batatas [J]. Acta Bot Boreali Occidentalia Sin, 2020, 40(4): 581-587. | |
| [21] | 韩俊杰, 王萍, 孙淑荣, 等. 气候变化对黑龙江紫丁香物候期的影响 [J]. 中国农学通报, 2023, 39(26): 130-136. |
| Han JJ, Wang P, Sun SR, et al. Effects of climate change on phenology of lilac in Heilongjiang province [J]. Chin Agric Sci Bull, 2023, 39(26): 130-136. | |
| [22] | 范小峰, 刘秀丽, 卢晶. 紫丁香的组织培养和快速繁殖 [J]. 林业科技通讯, 2024(5): 51-54. |
| Fan XF, Liu XL, Lu J. Tissue culture and rapid propagation of Syringa oblata [J]. For Sci Technol, 2024(5): 51-54. | |
| [23] | 孙全航, 康璠, 何淼, 等. 镉胁迫对紫丁香种子萌发及幼苗生长的影响 [J]. 种子, 2024, 43(2): 88-94. |
| Sun QH, Kang F, He M, et al. Effects of cadmium stress on seed germination and seedling growth of Syringa oblata L [J]. Seed, 2024, 43(2): 88-94. | |
| [24] | 王蕊, 王羽, 李彦慧, 等. 华北紫丁香肉桂酸4-羟化酶(SoC4H)基因的克隆及表达分析 [J]. 分子植物育种, 2016, 14(8): 2025-2030. |
| Wang R, Wang Y, Li YH, et al. Cloning and expression analysis of cinnamic acid 4-hydroxylase gene from Syringa oblata lindl [J]. Mol Plant Breed, 2016, 14(8): 2025-2030. | |
| [25] | 王蕊, 李彦慧, 郑健. 紫丁香查尔酮异构酶基因SoCHI的克隆及表达分析 [J]. 植物资源与环境学报, 2018, 27(3): 11-17. |
| Wang R, Li YH, Zheng J. Cloning and expression analysis on Chalcone isomerase gene SoCHI in Syringa oblata [J]. J Plant Resour Environ, 2018, 27(3): 11-17. | |
| [26] | 李文芳. 苹果4CL3基因启动子的克隆及其在花青素合成中的功能分析[D]. 兰州: 甘肃农业大学, 2023. |
| Li YF. Cloning of 4CL3 gene promoter and functional analysis of its role in anthocyanin synthesis in Malus domestica [D]. Lanzhou: Gansu Agricultural University, 2023. | |
| [27] | 马馨馨, 许洋, 赵欢欢, 等. 番茄4CL基因家族鉴定和氮素处理下的表达分析 [J]. 生物技术通报, 2022, 38(4): 163-173. |
| Ma XX, Xu Y, Zhao HH, et al. Identification of tomato 4CL gene family and expression analysis under nitrogen treatment [J]. Biotechnol Bull, 2022, 38(4): 163-173. | |
| [28] | Wang Y, Dou Y, Wang R, et al. Molecular characterization and functional analysis of chalcone synthase from Syringa oblata Lindl. in the flavonoid biosynthetic pathway [J]. Gene, 2017, 635: 16-23. |
| [29] | 马波, 李雷, 胡增辉, 等. 紫丁香SoF3′H对花瓣中花青苷合成的功能解析 [J]. 园艺学报, 2024, 51(8): 1833-1843. |
| Ma B, Li L, Hu ZH, et al. Functional analysis of SoF3′H in the synthesis of anthocyanidin in Syringa oblata petals [J]. Acta Hortic Sin, 2024, 51(8): 1833-1843. | |
| [30] | Uhlmann A, Ebel J. Molecular cloning and expression of 4-coumarate: coenzyme A ligase, an enzyme involved in the resistance response of soybean (Glycine max L.) against pathogen attack [J]. Plant Physiol, 1993, 102(4): 1147-1156. |
| [31] | 钟秀来, 赵倩, 朱顺华, 等. 芹菜Ag4CL1基因克隆及功能分析 [J/OL]. 分子植物育种, 2023. . |
| Zhong XL, Zhao Q, Zhu SH, et al. Cloning and expression analysis of Ag4CL1 gene in Apium graveolens [J/OL]. Mol Plant Breed, 2023. . | |
| [32] | 余珠, 欧阳乐军, 赵永国, 等. 4-香豆酸辅酶A连接酶(4CL)的研究进展 [J/OL]. 分子植物育种, 2023. . |
| Yu Z, Ouyang LJ, Zhao YG, et al. Research progress of 4-coumaric acid coenzyme aligase (4CL) [J/OL]. Mol Plant Breed, 2023. . | |
| [33] | Cao Y, Hu SL, Huang SX, et al. Molecular cloning, expression pattern, and putative cis-acting elements of a 4-coumarate: CoA ligase gene in bamboo (Neosinocalamus affinis) [J]. Electron J Biotechnol, 2012, 15(5): 9. |
| [34] | Allina SM, Pri-Hadash A, Theilmann DA, et al. 4-Coumarate: coenzyme A ligase in hybrid poplar. Properties of native enzymes, cDNA cloning, and analysis of recombinant enzymes [J]. Plant Physiol, 1998, 116(2): 743-754. |
| [35] | Yan C, Li CL, Jiang MC, et al. Systematic characterization of gene families and functional analysis of PvRAS3 and PvRAS4 involved in rosmarinic acid biosynthesis in Prunella vulgaris [J]. Front Plant Sci, 2024, 15: 1374912. |
| [36] | 田晓明, 颜立红, 向光锋, 等. 植物4香豆酸: 辅酶A连接酶研究进展 [J]. 生物技术通报, 2017, 33(4): 19-26. |
| Tian XM, Yan LH, Xiang GF, et al. Research progress on 4-coumarate: coenzyme a ligase (4CL) in plants [J]. Biotechnol Bull, 2017, 33(4): 19-26. | |
| [37] | 王浩哲, 钱海珍, 尤红艳, 等. 平原红猕猴桃花青苷积累规律及合成代谢关键基因发掘 [J]. 果树学报, 2024, 41(8): 1534-1545. |
| Wang HZ, Qian HZ, You HY, et al. Anthocyanin accumulation and related anabolic key genes in fruit of Pingyuanhong kiwifruit (Actinidia chinensis) [J]. J Fruit Sci, 2024, 41(8): 1534-1545. | |
| [38] | 母洪娜, 孙陶泽, 徐晨, 等. 桂花(Osmanthus fragrans Lour.)4 -香豆酸辅酶A连接酶(4CL)基因克隆与表达分析 [J]. 分子植物育种, 2016, 14(3): 536-541. |
| Mu HN, Sun TZ, Xu C, et al. Gene clone and expression analysis of 4-coumarate-Co a ligase in sweet Osmanthus (Osmanthus fragrans Lour.) [J]. Mol Plant Breed, 2016, 14(3): 536-541. | |
| [39] | Endler A, Martens S, Wellmann F, et al. Unusually divergent 4-coumarate: CoA-ligases from Ruta graveolens L [J]. Plant Mol Biol, 2008, 67(4): 335-346. |
| [40] | 唐亚琴. 油橄榄黄酮和木质素合成相关酶基因4CL的克隆及表达分析 [D]. 雅安: 四川农业大学, 2019. |
| Tang YQ. Cloning and characterization of the flavonoid and lignin synthesis related gene 4CL in olive [D]. Ya’an: Sichuan Agricultural University, 2019. | |
| [41] | Lee SH, Park YJ, Park SU, et al. Expression of genes related to phenylpropanoid biosynthesis in different organs of Ixeris dentata var. albiflora [J]. Molecules, 2017, 22(6): 901. |
| [42] | 潘翔. 毛白杨4CL基因启动子及APX的结构与功能研究 [D]. 北京: 北京林业大学, 2013. |
| Pan X. Study on the structure and function of the 4CL gene promoters and APX in Populus tomentosa [D]. Beijing: Beijing Forestry University, 2013. | |
| [43] | Laoué J, Fernandez C, Ormeño E. Plant flavonoids in mediterranean species: a focus on flavonols as protective metabolites under climate stress [J]. Plants, 2022, 11(2): 172. |
| [44] | Wang JX, Wang X, Ma B, et al. SoNAC72-SoMYB44/SobHLH130 module contributes to flower color fading via regulating anthocyanin biosynthesis by directly binding to the SoUFGT1 promoter in lilac (Syringa oblata) [J]. Hortic Res, 2025, 12(3): uhae326. |
| [45] | 刘淑华, 臧丹丹, 孙燕, 等. 花青素生物合成途径及关键酶研究进展 [J]. 土壤与作物, 2022, 11(3): 336-346. |
| Liu SH, Zang DD, Sun Y, et al. Research advances on biosynthesis pathway of anthocyanins and relevant key enzymes [J]. Soil Crop, 2022, 11(3): 336-346. | |
| [46] | 刘国元, 方威, 余春梅, 等. 花青素调控植物花色的研究进展 [J]. 安徽农业科学, 2021, 49(3): 1-4, 9. |
| Liu GY, Fang W, Yu CM, et al. Research progress on anthocyanins regulated plant flower color [J]. J Anhui Agric Sci, 2021, 49(3): 1-4, 9. | |
| [47] | 彭贞贞, 钟传飞, 王宝刚, 等. ‘红颜’草莓果实成熟过程中花色苷积累及合成途径基因表达的分析 [J]. 食品工业科技, 2023, 44(14): 346-354. |
| Peng ZZ, Zhong CF, Wang BG, et al. Analysis of anthocyanin accumulation and gene expression of anthocyanin synthesis pathway during fruit ripening of ‘benihoppe’ strawberry [J]. Sci Technol Food Ind, 2023, 44(14): 346-354. |
| [1] | LYU Cheng-cong, HENG Meng, CHEN Si-qi, JIN Xue-hua. Cloning and Functional Analysis of ZhGSTF Related to Anthocyanin Transport Zantedeschia hybrida [J]. Biotechnology Bulletin, 2026, 42(1): 161-169. |
| [2] | CHEN Jing-huan, FANG Guo-nan, ZHU Wen-hao, YE Guang-ji, SU Wang, HE Miao-miao, YANG Sheng-long, ZHOU Yun. Starch Characterization and Related Gene Expression Analysis of Potato Germplasm Resources [J]. Biotechnology Bulletin, 2026, 42(1): 170-183. |
| [3] | DONG Xiang-xiang, MIAO Bai-ling, XU He-juan, CHEN Juan-juan, LI Liang-jie, GONG Shou-fu, ZHU Qing-song. Bioinformatics Analysis and Flowering Regulation Function of FveBBX20 Gene in Woodland Strawberry [J]. Biotechnology Bulletin, 2025, 41(9): 115-123. |
| [4] | ZHANG Yong-yan, GUO Si-jian, LI Jing, HAO Si-yi, LI Rui-de, LIU Jia-peng, CHENG Chun-zhen. Gene Cloning and Functional Analysis of the Anthocyanin-related VcGSTF19 Gene in Blueberry (Vaccinium corymbosum L.) [J]. Biotechnology Bulletin, 2025, 41(9): 139-146. |
| [5] | CHEN Qiang, YU Ying-fei, ZHANG Ying, ZHANG Chong. Regulatory Effect of Methyl Jasmonate on Postharvest Chilling Injury in Oriental Melon ‘Emerald’ [J]. Biotechnology Bulletin, 2025, 41(9): 105-114. |
| [6] | XU Xiao-ping, YANG Cheng-long, HE Xing, GUO Wen-jie, WU Jian, FANG Shao-zhong. Cloning of the LoAPS1 and Its Function Analysis during the Process of Dormancy Release in Lilium [J]. Biotechnology Bulletin, 2025, 41(9): 195-206. |
| [7] | CHENG Xue, FU Ying, CHAI Xiao-jiao, WANG Hong-yan, DENG Xin. Identification of LHC Gene Family in Setaria italica and Expression Analysis under Abiotic Stresses [J]. Biotechnology Bulletin, 2025, 41(8): 102-114. |
| [8] | LI Kai-jie, WU Yao, LI Dan-dan. Cloning of Gene CtbHLH128 in Safflower and Response Function Regulating Drought Stress [J]. Biotechnology Bulletin, 2025, 41(8): 234-241. |
| [9] | REN Rui-bin, SI Er-jing, WAN Guang-you, WANG Jun-cheng, YAO Li-rong, ZHANG Hong, MA Xiao-le, LI Bao-chun, WANG Hua-jun, MENG Ya-xiong. Identification and Expression Analysis of GH17 Gene Family of Pyrenophora graminea [J]. Biotechnology Bulletin, 2025, 41(8): 146-154. |
| [10] | KANG Qin, WANG Xia, SHEN Ming-yang, XU Jing-tian, CHEN Shi-lan, LIAO Ping-yang, XU Wen-zhi, WU Wei, XU Dong-bei. Cloning and Expression Analysis of the UV-B Receptor Gene McUVR8 in Mentha canadensis L. [J]. Biotechnology Bulletin, 2025, 41(8): 255-266. |
| [11] | ZENG Dan, HUANG Yuan, WANG Jian, ZHANG Yan, LIU Qing-xia, GU Rong-hui, SUN Qing-wen, CHEN Hong-yu. Genome-wide Identification and Expression Analysis of bZIP Transcription Factor Family in Dendrobium officinale [J]. Biotechnology Bulletin, 2025, 41(8): 197-210. |
| [12] | HAN Yi, HOU Chang-lin, TANG Lu, SUN Lu, XIE Xiao-dong, LIANG Chen, CHEN Xiao-qiang. Cloning and Preliminary Functional Analysis of HvERECTA Gene in Hordeum vulgare [J]. Biotechnology Bulletin, 2025, 41(7): 106-116. |
| [13] | WEI Yu-jia, LI Yan, KANG Yu-han, GONG Xiao-nan, DU Min, TU Lan, SHI Peng, YU Zi-han, SUN Yan, ZHANG Kun. Cloning and Expression Analysis of the CrMYB4 Gene in Carex rigescens [J]. Biotechnology Bulletin, 2025, 41(7): 248-260. |
| [14] | XU Hui-zhen, SHANTWANA Ghimire, RAJU Kharel, YUE Yun, SI Huai-jun, TANG Xun. Analysis of the Potato SUMO E3 Ligase Gene Family and Cloning and Expression Pattern of StSIZ1 [J]. Biotechnology Bulletin, 2025, 41(6): 119-129. |
| [15] | PEI Jing-qi, ZHAO Meng-ran, HUANG Chen-yang, WU Xiang-li. Discovery and Verification of a Functional Gene Influencing the Growth and Development of Pleurotus ostreatus [J]. Biotechnology Bulletin, 2025, 41(6): 327-334. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||