








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (4): 96-106.doi: 10.13560/j.cnki.biotech.bull.1985.2020-0887
Previous Articles Next Articles
ZHANG Yao-xin( ), WANG Liang-jie, ZHENG Wen, XU Han-qin, ZHENG Lian, ZHONG Jing(
), WANG Liang-jie, ZHENG Wen, XU Han-qin, ZHENG Lian, ZHONG Jing( )
)
Received:2020-07-16
Online:2021-04-26
Published:2021-05-13
Contact:
ZHONG Jing
E-mail:helen20041986@sina.com;jjing2003_1@163.com
ZHANG Yao-xin, WANG Liang-jie, ZHENG Wen, XU Han-qin, ZHENG Lian, ZHONG Jing. Study on Enzyme Production of a Chitinase-producing Strain Achromobacter sp. ZWW8 by Fermentation and Its Enzymatic Characterization[J]. Biotechnology Bulletin, 2021, 37(4): 96-106.
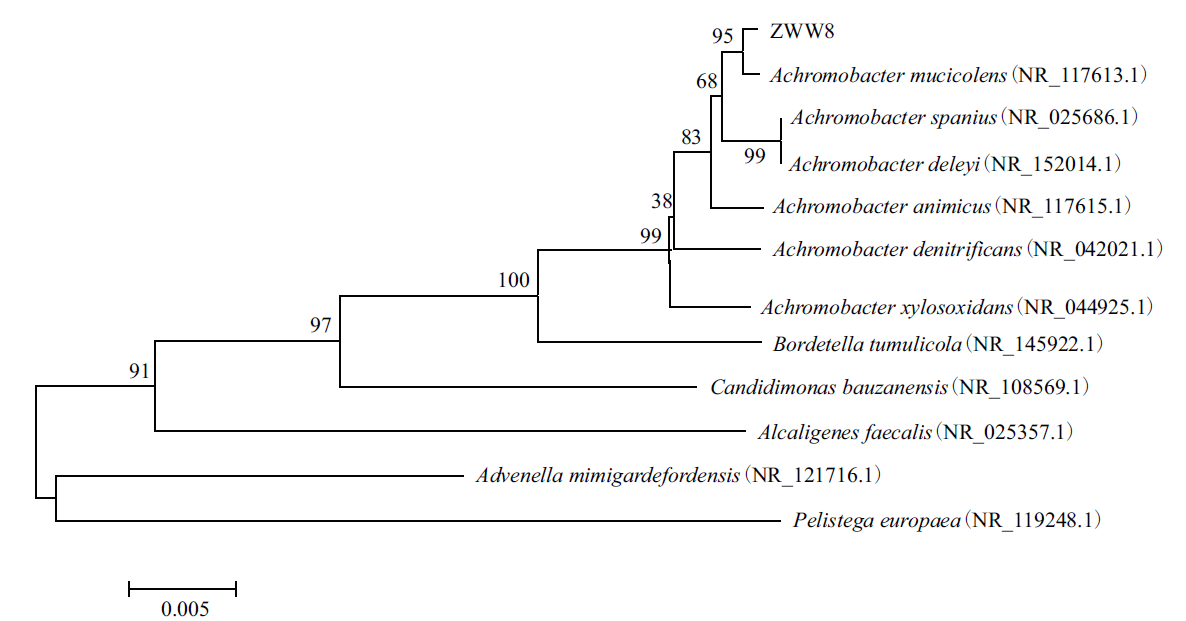
Figure 1 Phylogenetic tree of strain ZWW8 based on 16S rDNA sequences Numbers in parentheses refer to the sequence’ accession number in GenBank. The number at each branch point is the percentage supported by bootstrap. Bar: 0.5% sequence divergence
| 碳源Carbon source | 酶活Enzyme activity/(U·L-1) |
|---|---|
| 葡萄糖 Glucose | 0.2±0.8 |
| 胶体几丁质 Colloidal chitin | 20.0±1.9 |
| 粉末几丁质 Chitin powder | 4.2±0.3 |
| 胶体壳聚糖 Colloidal chitosan | 0.0±0.1 |
| 水溶性壳聚糖 Soluble chitosan | 0.2±0.1 |
| N-乙酰氨基葡萄糖 N-Acetyl-D-glucosamine | 0.4±0.6 |
Table 1 Effect of different carbon sources on chitinase production by Achromobacter sp. ZWW8
| 碳源Carbon source | 酶活Enzyme activity/(U·L-1) |
|---|---|
| 葡萄糖 Glucose | 0.2±0.8 |
| 胶体几丁质 Colloidal chitin | 20.0±1.9 |
| 粉末几丁质 Chitin powder | 4.2±0.3 |
| 胶体壳聚糖 Colloidal chitosan | 0.0±0.1 |
| 水溶性壳聚糖 Soluble chitosan | 0.2±0.1 |
| N-乙酰氨基葡萄糖 N-Acetyl-D-glucosamine | 0.4±0.6 |
| 氮源Nitrogen source | 酶活Enzyme activity(U·L-1) |
|---|---|
| 氯化铵 Ammonium chloride | 20.7±1.3 |
| 硫酸铵 Ammonium sulfate | 17.8±1.5 |
| 硝酸钠 Sodium nitrate | 6.4±1.9 |
| 尿素 Urea | 13.3±1.0 |
| 蛋白胨 Peptone | 12.1±1.8 |
| 硝酸铵Ammonium nitrate | 19.2±0.2 |
Table2 Effect of different nitrogen sources on chitinase production by Achromobacter sp. ZWW8
| 氮源Nitrogen source | 酶活Enzyme activity(U·L-1) |
|---|---|
| 氯化铵 Ammonium chloride | 20.7±1.3 |
| 硫酸铵 Ammonium sulfate | 17.8±1.5 |
| 硝酸钠 Sodium nitrate | 6.4±1.9 |
| 尿素 Urea | 13.3±1.0 |
| 蛋白胨 Peptone | 12.1±1.8 |
| 硝酸铵Ammonium nitrate | 19.2±0.2 |
| 金属离子Metal ion | 相对酶活Relative activity/% |
|---|---|
| Hg2+ | 3.1±3.3 |
| Zn2+ | 84.4±8.5 |
| Mg2+ | 96.1±7.1 |
| Ca2+ | 103.9±9.3 |
| Ag+ | 99.2±6.8 |
| Cu2+ | 75.9±0.1 |
| Ni2+ | 114.7±10.3 |
Table 3 Effect of metal ions on chitinase activity by strain ZWW8
| 金属离子Metal ion | 相对酶活Relative activity/% |
|---|---|
| Hg2+ | 3.1±3.3 |
| Zn2+ | 84.4±8.5 |
| Mg2+ | 96.1±7.1 |
| Ca2+ | 103.9±9.3 |
| Ag+ | 99.2±6.8 |
| Cu2+ | 75.9±0.1 |
| Ni2+ | 114.7±10.3 |

Fig.11 SDS-PAGE analysis of strain ZWW8 chitinase purified by chitin affinity chromatography M: Low molecular weight marker. 1: Crude chitinase. 2: Effluent. 3,4: Scrub solution, 5: Purified chitinase
| [1] |
Kurita K. Chemistry and application of chitin and chitosan[J]. Polymer Degradation and Stability, 1998,59(1):117-120.
doi: 10.1016/S0141-3910(97)00160-2 URL |
| [2] | 欧阳石文, 冯兰香, 赵开军. 几丁质酶的三级结构和催化机制[J]. 生命的化学, 2001,21(2):131-133. |
| Ou-Yang SW, Feng LX, Zhao KJ. The tertiary structure and catalytic mechanism of chitinase[J]. Chemistry of Life, 2001,21(2):131-133. | |
| [3] |
Bhattacharya D, Nagpure A, Gupta RK. Bacterial chitinases:properties and potential[J]. Critical Reviews in Biotechnology, 2007,27(1):21-28.
doi: 10.1080/07388550601168223 URL |
| [4] |
Nagpure A, Bharti , Gupta RK. Chitinases:in agriculture and human healthcare[J]. Critical Reviews in Biotechnology, 2013,34(3):215-232.
doi: 10.3109/07388551.2013.790874 URL |
| [5] | Dahiya N, Tewari R, Hoondal GS. Biotechnological aspects of chitinolytic enzymes:a review[J]. Applied Microbiology & Biotechnology, 2006,71(6):773-782. |
| [6] | 郭玉莲. 微生物几丁质酶及其在植物病害防治中的作用[J]. 中国农学通报, 2005(1):283-285. |
| Guo Y. Chitinase and its effects on plant disease prevention[J]. Chinese Agricultural Science Bulletin, 2005(1):283-285. | |
| [7] | 黄乾生, 谢晓兰, 陈清西. 几丁质酶的结构特征与功能[J]. 厦门大学学报:自然科学版, 2008,47(z2). 232-235. |
| Huang QS, Xie XL, Chen QX. The structure and function of chitinase[J]. Journal of Xiamen University:Natural Science, 2008,47(z2). 232-235 . | |
| [8] | 冯金荣, 惠丰立, 文祯中. 真菌几丁质酶及其在植物真菌病害防治中的作用[J]. 河南农业科学, 2006(8):83-86. |
| Feng JR, Hui FL, Wen ZZ. Fungal chitinase and its role in the control of plant fungal diseases[J]. Journal of Henan Agricultural Sciences, 2006(8):83-86. | |
| [9] | Sakuda S, Isogai A, Makita T, et al. Structures of allosamidins, novel insect chitinase inhibitors, produced by actinomycetes[J]. Journal of the Agricultural Chemical Society of Japan, 2006,51(12):3251-3259. |
| [10] |
Liaqat F, Eltem R. Chitooligosaccharides and their biological activities:A comprehensive review[J]. Carbohydrate Polymers, 2018,184:243-259.
doi: 10.1016/j.carbpol.2017.12.067 URL |
| [11] |
Chigaleĭchik AG, Pirieva DA, Rylkin SS, et al. Biosynjournal of chitinase by Achromobacter liquefaciens[J]. Mikrobiologiia, 1976,45:475.
pmid: 12452 |
| [12] | Takashi T, Hideyuki M. Antibacterial, anti-nematode and/or plant-cell activating composition,chitinolytic microorganisms for producing the same:European, EP0401560[P]. 1993-05-04. |
| [13] | Bholay AD, Bhushan K, Rameez S, et al. Novel chitinolytic potential of Achromobacter denitrificans isolated from fishery waste[J]. IOSR Journal of Pharmacy and Biological Sciences, 2015,10(4):31-36. |
| [14] |
Baker GC, Smith JJ, Cowan DA . Review and re-analysis of domain-specific 16S primers[J]. Journal of Microbiological Methods, 2004,55(3):541-555.
doi: 10.1016/j.mimet.2003.08.009 URL |
| [15] | 张军毅, 朱冰川, 徐超, 等. 基于分子标记的宏基因组16S rRNA基因高变区选择策略[J]. 应用生态学报, 2015,26(11):3545-3553. |
| Zhang JY, Zhu BC, Xu C, et al. Strategy of selecting 16S rRNA hypervariable regions for matagenome-phylogenetic marker genes based analysis[J]. Chinese Journal of Applied Ecology, 2015,26(11):3545-3553. | |
| [16] |
Kim SK, Rajapakse N. Enzymatic production and biological activities of chitosan oligosaccharides(COS):A review[J]. Carbohydrate Polymers, 2005,62(4):357-368.
doi: 10.1016/j.carbpol.2005.08.012 URL |
| [17] |
Dou J, Xu Q, Tan C, et al. Effects of chitosan oligosaccharides on neutrophils from glycogen-induced peritonitis mice model[J]. Carbohydrate Polymers, 2009,75(1):119-124.
doi: 10.1016/j.carbpol.2008.07.005 URL |
| [18] |
Aam BB, Heggset EB, Norberg AL, et al. Production of chitooligosaccharides and their potential applications in medicine[J]. Marine Drugs, 2010,8(5):1482-1517.
doi: 10.3390/md8051482 URL |
| [19] |
Hough DW, Danson MJ. Extremozymes[J]. Current Opinion in Chemical Biology, 1999,3(1):39-46.
pmid: 10021406 |
| [20] |
Karan R, Capes MD, Dassarma S . Function and biotechnology of extremophilic enzymes in low water activity[J]. Aquatic Biosystems, 2012,8(1):1-4.
doi: 10.1186/2046-9063-8-1 URL |
| [21] |
Vaidya R, Shah I, Vyas P, et al. Production of chitinase and its optimization from a novel isolate Alcaligenes xylosoxydans:potential in antifungal biocontrol[J]. World Journal of Microbiology & Biotechnology, 2001,17(7):691-696.
doi: 10.1023/A:1012927116756 URL |
| [22] | Vaidya R, Vyas P, Chhatpar HS. Statistical optimization of medium components for the production of chitinase by Alcaligenes xylosoxydans[J]. Enzyme & Microbial Technology, 2003,33(1):92-96. |
| [23] |
Vaidya R, Roy S, Macmil S, et al. Purification and characterization of chitinase from Alcaligenes xylosoxydans[J]. Biotechnology Letters, 2003,25(9):715-717.
doi: 10.1023/A:1023406630791 URL |
| [24] | 付星, 闫巧娟, 江正强, 等. 高产几丁质酶巴伦葛兹类芽孢杆菌的筛选和发酵条件优化[J]. 微生物学通报, 2015,42(4):625-633. |
| Fu X, Yan QJ, Jiang ZQ, et al. Screening of a high-level chitinase producing strain, Paenibacillus barengoltzii and optimization of its fermentation conditions[J]. Microbiology China, 2015,42(4):625-633. | |
| [25] |
Vyas P, Deshpande MV . Chitinase production by Myrothecium verrucaria and its significance for fungal mycelia degradation[J]. J Gen Appl Microbiol, 1989,35(5):343-350.
doi: 10.2323/jgam.35.343 URL |
| [26] |
Kavitha A, Vijaylakshmi M. Partial purification and antifungal profile of chitinase produced by Streptomyces tendae TK-VL_333[J]. Annals of Microbiology, 2011,61(3):597-603.
doi: 10.1007/s13213-010-0178-1 URL |
| [27] |
Uchiyama T, Katouno F, Nikaidou N, et al. Roles of the exposed aromatic residues in crystalline chitin hydrolysis by chitinase A from Serratia marcescens 2170[J]. Journal of Biological Chemistry, 2001,276(44):41343-41349.
doi: 10.1074/jbc.M103610200 URL |
| [28] |
Orikoshi H, Nakayama S, Miyamoto K, et al. Roles of four chitinases(ChiA, ChiB, ChiC, and ChiD)in the chitin degradation system of marine bacterium Alteromonas sp. strain O-7[J]. Applied and Environmental Microbiology, 2005,71(4):1811-1815.
doi: 10.1128/AEM.71.4.1811-1815.2005 URL |
| [1] | CHENG Ya-nan, ZHANG Wen-cong, ZHOU Yuan, SUN Xue, LI Yu, LI Qing-gang. Synthetic Pathway Construction of Producing 2'-fucosyllactose by Lactococcus lactis and Optimization of Fermentation Medium [J]. Biotechnology Bulletin, 2023, 39(9): 84-96. |
| [2] | YANG Jun-zhao, ZHANG Xin-rui, ZHAO Guo-zhu, ZHENG Fei. Structure and Function Analysis of Novel GH5 Multi-domain Cellulase [J]. Biotechnology Bulletin, 2023, 39(4): 71-80. |
| [3] | LI Ji-hong, JING Yu-ling, MA Gui-zhen, GUO Rong-jun, LI Shi-dong. Genome Construction of Achromobacter 77 and Its Characteristics on Chemotaxis and Antibiotic Resistance [J]. Biotechnology Bulletin, 2022, 38(9): 136-146. |
| [4] | WANG Yu-chen, DING Zun-dan, GUAN Fei-fei, TIAN Jian, LIU Guo-an, WU Ning-feng. Identification of the Thermostable Laccase Gene ba4 and Characterization of Its Enzymatic Properties [J]. Biotechnology Bulletin, 2022, 38(8): 252-260. |
| [5] | FU Qiao, LIN Qi-lan, XUE Qiang, XIONG Hai-rong, WANG Ya-wei. Effects of CBM41 N-terminal Truncation on the Enzymological Properties of the Pullulanase from Bacillus subtilis 168 [J]. Biotechnology Bulletin, 2022, 38(6): 245-251. |
| [6] | WANG Xiao-tao, ZOU Hang, WU Yi, XIANG Shen-wei, LV Hua, LIU Chao-lan, LIN Jia-fu, WANG Xin-rong, CHU Yi-wen, SONG Tao. Heterologous Expression and Enzymatic Properties Analysis of Novel β-agarase Aga2 from Paraglaciecola hydrolytica [J]. Biotechnology Bulletin, 2022, 38(11): 258-268. |
| [7] | ZHOU Jing, HUANG Wen-mao, QIN Li-jun, HAN Li-zhen. Construction of Mixed Fermentation System of Four PGPR Strains and Evaluation of Its Promoting Effect [J]. Biotechnology Bulletin, 2021, 37(4): 116-126. |
| [8] | WU Rong, CAO Jia-rui, CAO Jun, LIU Fei-xiang, YANG Meng, SU Er-zheng. Expression and Fermentation Optimization of Candida antarctica Lipase B in Escherichia coli [J]. Biotechnology Bulletin, 2021, 37(2): 138-148. |
| [9] | PAN Jing, HUANG Cui-hua, PENG Fei, YOU Quan-gang, LIU Fei-yao, XUE Xian. Mechanisms of Salt Tolerance and Growth Promotion in Plant Induced by Plant Growth-Promoting Rhizobacteria [J]. Biotechnology Bulletin, 2020, 36(9): 75-87. |
| [10] | LI Si-qi, YANG Jing-dan, LIU Lin, LIU Er-jia, WANG Xiao-hui. Denitrification Characteristics of Aerobic Denitrifying Bacteria Achromobacter sp. L16 [J]. Biotechnology Bulletin, 2020, 36(6): 93-101. |
| [11] | YUE Li-xiao, LI Deng-yun, ZHANG Jing-jing, TONG Lei. Isolation and Application Potential Exploration of a Diuron-degrading Bacterium [J]. Biotechnology Bulletin, 2020, 36(6): 110-119. |
| [12] | WANG Hui-lan, WU Jin-yong, CHEN Xiang-song, YUAN Li-xia, ZHU Wei-wei, YAO Jian-ming. Immobilization of N-acetylneuraminic Acid Aldolaseand Properties of the Immobilized Enzyme [J]. Biotechnology Bulletin, 2020, 36(6): 165-173. |
| [13] | WANG Zhuo, LUO Xue-gang, DING Han-lin, YANG Hao. Isolation and Identification of a Uranium-resistant Strain and Effect of Its Characteristics on Growth Promoting [J]. Biotechnology Bulletin, 2019, 35(1): 42-50. |
| [14] | LI Cheng-ming, WANG Jia-yi, LU Lei. Extracellular Expression of Laccase Gene from Bacillus pumilus LC01 in Pichia pastoris and Characterization of the Recombinant Laccase [J]. Biotechnology Bulletin, 2018, 34(3): 185-193. |
| [15] | GUO Ji-ping, MA Guang, WANG Zhi-jie, QI Shan-hou, WANG Bao-mei, SU Chang-qing. Identification and Analysis of Biocontrol Proteins from a Strain of Bacillus amyloliquefaciens [J]. Biotechnology Bulletin, 2018, 34(1): 202-207. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||