








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (5): 98-107.doi: 10.13560/j.cnki.biotech.bull.1985.2020-1149
Previous Articles Next Articles
BAI Fu-mei1( ), LI Zhi-min2, WANG Xiao-qin1, HU Zi-wei1, BAO Ling-ling1, LI Zhi-min1,3(
), LI Zhi-min2, WANG Xiao-qin1, HU Zi-wei1, BAO Ling-ling1, LI Zhi-min1,3( )
)
Received:2020-09-09
Online:2021-05-26
Published:2021-06-11
Contact:
LI Zhi-min
E-mail:bfm18270860135@163.com;zmlizm@126.com
BAI Fu-mei, LI Zhi-min, WANG Xiao-qin, HU Zi-wei, BAO Ling-ling, LI Zhi-min. Biochemical Characterization and Structural Analysis of N-acetylornithine Transaminase from Synechocystis sp. PCC6803[J]. Biotechnology Bulletin, 2021, 37(5): 98-107.
| Primer | Primer sequence(5'-3') | Restriction enzyme |
|---|---|---|
| slr1022-F | GGAATTCCATATGACCTATTCC-CCTGTTGTTGAATC | Nde I |
| slr1022-R | CCGCTCGAGTCAAACCAAAGT-GGCGATCGCCTGAC | Xho I |
Table 1 PCR primer sequences
| Primer | Primer sequence(5'-3') | Restriction enzyme |
|---|---|---|
| slr1022-F | GGAATTCCATATGACCTATTCC-CCTGTTGTTGAATC | Nde I |
| slr1022-R | CCGCTCGAGTCAAACCAAAGT-GGCGATCGCCTGAC | Xho I |
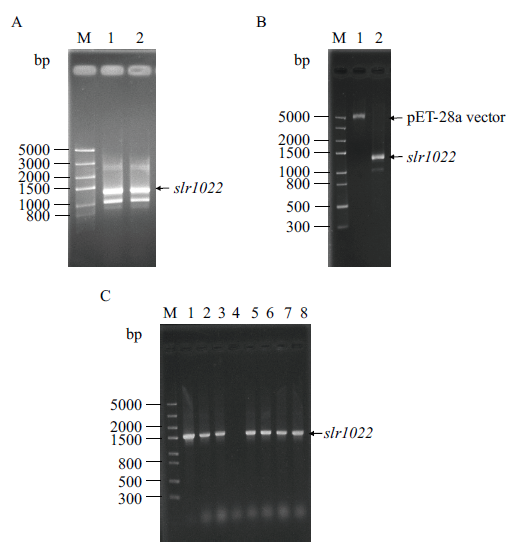
Fig. 1 Cloning of slr1022 gene from Synechocystis sp. PCC6803 and construction of its recombinant plasmid pET28a-slr1022 A: PCR amplification product of slr1022 gene. M: DNA marker. Lane 1-2: PCR product. B: Double enzymatic digestion products of slr1022 gene and pET-28a vector. M: DNA marker. Lane 1-2: Double enzymatic digestion product of pET-28a vector and slr1022 gene, respectively. C: Identification of recombinant plasmid pET28a-slr1022. M: DNA marker. Lane 1: Positive control. Lane 2-8: Colony PCR product

Fig. 2 Expression of recombinant Slr1022 protein M: Protein marker. Lane 1-3: Expression of pET-28a plasmid (cell lysate, supernatant and pellet, respectively). Lane 4-6: Expression of pET28a-slr1022 recombinant plasmid (cell lysate, supernatant and pellet, respectively)
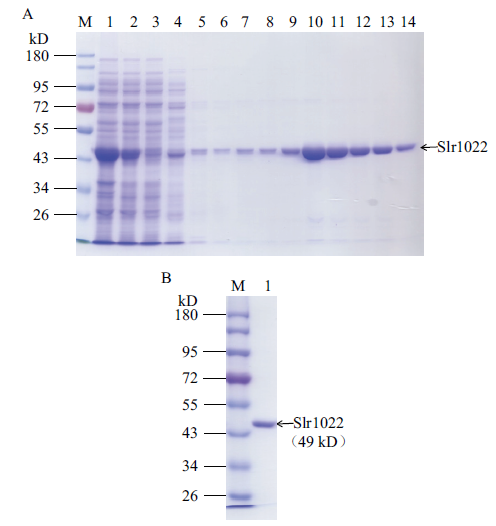
Fig. 3 SDS-PAGE of recombinant Slr1022 protein A: Purification of recombinant Slr1022 protein. M: Protein marker. Lane 1-3: Cell lysate, supernatant and flow through. Lane 4-8: 20, 40, 60, 80, 100 mmol/L imidazole elution. 9-14: 200 mmol/L imidazole elution. B: The purified Slr1022 protein. M: Protein marker. Lane 1: The purified Slr1022 protein
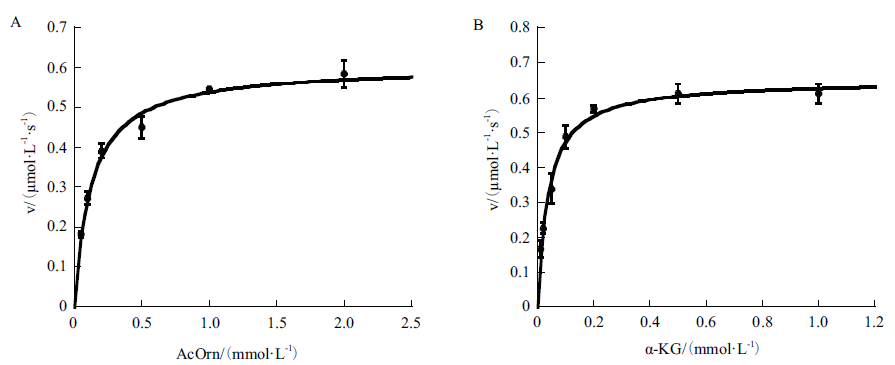
Fig. 4 Kinetic profiles of recombinant Slr1022 protein A: Plot of the initial velocities as function of N-acetylornithine concentrations, the concentration of α-ketoglutarate was fixed at 1 mmol/L. B: Plot of the initial velocities as function of α-ketoglutarate concentrations, the concentration of N-acetylornithine was fixed at 2 mmol/L. Data are expressed as the mean ± standard deviation, n = 3

Fig. 6 Protein sequences alignment of N-acetylornithine aminotransferases from different sources slr1022, 2E54, 6W7X, 2EH6 and 2PB0 were N-acetylornithine aminotransferases from Synechocystis PCC6803, Thermotoga maritima, Stenotrophomonas maltophilia, Aquifex aeolicus and Salmonella typhimurium respectively. Residues interacting with PLP are marked by▲, and★ indicates the residues forming Schiff base with PLP and ◆ indicates the residues from the other subunit which interact with PLP

Fig. 7 Homologous modeling structure of Slr1022 protein A: Cartoon structure of Slr1022 protein. B: Enlarged schematic diagram of active site. C: The conserved amino acid residues in the active site. The colors of α-helix, β-strands and loops are depicted as red, yellow and green, respectively. The cofactor PLP and the conserved amino acid residues are shown as sticks, and their carbon atoms are light blue and green, respectively. The yellow dotted lines indicate the hydrogen bonds interacting with PLP. The modeled structure is depicted by SWISS-MODEL based on the structure of N-acetylornithine aminotransferase of Thermotoga maritima (PBD: 2E54)
| [1] | Burkill PH, Leakey RJG, Owens NJP, et al. Synechococcus and its importance to the microbial foodweb of the northwestern Indian Ocean[J]. Deep-Sea Research II, 1993,40(3):773-782. |
| [2] | 裴广胜. 模式蓝细菌集胞藻中分子调控系统的解析[D]. 天津:天津大学, 2017. |
| Pei GS. Functions analysis of regulatory molecules in the model cyanobacterium Synechocystis sp. PCC 6803[D]. Tianjin:Tianjin University, 2017. | |
| [3] | Stanier RY, Kunisawa R, Mandel R, et al. Purification and properties of unicellular blue-green algae(order chroococcales)[J]. Bacterological Reviews, 1971,35(2):171-205. |
| [4] |
Kaneko T, Sato S, Kotani H, et al. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. sequence determination of the entire genome and assignment of potential protein-coding regions(Supplement)[J]. DNA Research, 1996,3(3):185-209.
pmid: 8905238 |
| [5] | Gao Z, Zhao H, Li Z, et al. Photosynthetic production of ethanol from carbon dioxide in genetically engineered cyanobacteria[J]. Energy & Environmental Science, 2012,5(12):9857-9865. |
| [6] |
Aizouq M, Peisker H, Gutbrod K, et al. Triacylglycerol and phytyl ester synjournal in Synechocystis sp. PCC6803[J]. PNAS, 2020,117(11):6216-6222.
doi: 10.1073/pnas.1915930117 URL |
| [7] | 张兵, 王利红, 徐玉新, 等. 集胞藻(Synechocystis sp. PCC6803)对砷吸收转化特性的初步研究[J]. 生态毒理学报, 2011,6(6):629-633. |
| Zhang B, Wang LH, Xu YX, et al. Study on absorption and transformation of arsenic in blue alga(Synechocystis sp. PCC6803)[J]. Asian J Ecotox, 2011,6(6):629-633. | |
| [8] |
Pearce J, Carr NG. The metabolism of acetate by the blue-green algae, Anabaena variabilis and Anacystis nidulans[J]. Journal of General Microbiology, 1967,49:301-313.
pmid: 6077932 |
| [9] |
Xiong W, Brune D, Vermaas WF. The γ-aminobutyric acid shunt contributes to closing the tricarboxylic acid cycle in Synechocystis sp. PCC 6803[J]. Mol Microbiol, 2014,93(4):786-796.
doi: 10.1111/mmi.12699 URL |
| [10] | Zhang S, Qian X, Chang S, et al. Natural and synthetic variants of the tricarboxylic acid cycle in cyanobacteria:introduction of the GABA shunt into Synechococcus sp. PCC 7002[J]. Frontiers in Microbiology, 2016,7:1972. |
| [11] |
Philipp C, Mehta PK. From cofactor to enzymes. The molecular evolution of pyridoxal-5’-phosphate-dependent enzymes[J]. The Chemical Record, 2001,1:436-447.
doi: 10.1002/(ISSN)1528-0691 URL |
| [12] |
Rajaram V, Ratna Prasuna P, Savithri HS, et al. Structure of biosynthetic N-acetylornithine aminotransferase from Salmonella typhimurium:studies on substrate specificity and inhibitor binding[J]. Proteins, 2008,70(2):429-441.
doi: 10.1002/prot.21567 URL |
| [13] |
Ledwidge R, Blanchard JS. The dual biosynthetic capability of N-acetylornithine aminotransferase in arginine and lysine biosynjournal[J]. Biochemistry, 1999,38(10):3019-3024.
doi: 10.1021/bi982574a URL |
| [14] | 王丹. 牛肝谷氨酸脱氢酶的分离纯化及酶学性质和功能基团研究[D]. 重庆:西南大学, 2016. |
| Wang D. Isolation, purification, properties and modification of groups of the glutamate dehydrogenase from bovine liver[D]. Chongqing:Southwest University, 2016. | |
| [15] | 徐美娟, 张显, 饶志明, 等. 钝齿棒杆菌N-乙酰鸟氨酸转氨酶的克隆表达分析及其重组菌的精氨酸发酵[J]. 生物工程学报, 2011,27(7):1013-1023. |
| Xu M, Zhang X, Rao Z, et al. Cloning, expression and characterization of N-acetylornithine aminotransferase from Corynebacterium crenatum and its effects on L-arginine fermentation[J]. Chinese J Biotechnol, 2011,27(7):1013-1023. | |
| [16] |
Friedrich B, Friedrich CG, Magasanik B. Catabolic N2-acetylornithine 5-aminotransferase of Klebsiella aerogenes:control of synjournal by induction, catabolite repression, and activation by glutamine synthetase[J]. J Bacteriol, 1978,133(2):686-691.
pmid: 24039 |
| [17] |
Mehta PK, Hale TI, Christen P. Aminotransferases:demonstration of homology and division into evolutionary subgroups[J]. European Journal of Biochemistry 1993,214:549-561.
pmid: 8513804 |
| [18] |
Shen BW, Hennig M, Hohenester E, et al. Crystal structure of human recombinant ornithine aminotransferase[J]. Journal of Molecular Biology, 1998,277:81-102.
doi: 10.1006/jmbi.1997.1583 URL |
| [19] |
Toney MD, Pascarella S, Biase DD. Active site model for γ-aminobutyrate aminotransferase explains substrate specificity and inhibitor reactivities[J]. Protein Science, 1995,4:2366-2374.
pmid: 8563634 |
| [20] |
Steinhauser D, Fernie AR, Araujo WL. Unusual cyanobacterial TCA cycles:not broken just different[J]. Trends in Plant Science, 2012,17(9):503-509.
doi: 10.1016/j.tplants.2012.05.005 pmid: 22658681 |
| [21] |
Fait A, Nesi AN, et al. Targeted enhancement of glutamate-to-gamma-aminobutyrate conversion in Arabidopsis seeds affects carbon-nitrogen balance and storage reserves in a development-dependent manner[J]. Plant Physiol, 2011,157:1026-1042.
doi: 10.1104/pp.111.179986 URL |
| [22] |
Bouche N, Fait A, Bouchez D, et al. Mitochondrial succinic-semialdehyde dehydrogenase of the γ-aminobutyrate shunt is required to restrict levels of reactive oxygen intermediates in plants[J]. PNAS, 2003,100(11):6843-6848.
doi: 10.1073/pnas.1037532100 URL |
| [23] |
Palanivelu R, Edlund AF, Brass L, et al. Pollen tube growth and guidance is regulated by POP2, an Arabidopsis gene that controls GABA levels[J]. Cell, 2003,114:47-59.
doi: 10.1016/S0092-8674(03)00479-3 URL |
| [24] | Decavel C, Van Den Pol AN. GABA:a dominant neurotransmitter in the hypothalamus[J]. The Journal of Comparam Neurologyneurology, 1990,302:1019-1037. |
| [25] |
Granger AJ, Wallace ML, Sabatini BL. Multi-transmitter neurons in the mammalian central nervous system[J]. Current Opinion in Neurobiology, 2017,45:85-91.
doi: S0959-4388(17)30016-8 pmid: 28500992 |
| [1] | MEI Huan, LI Yue, LIU Ke-meng, LIU Ji-hua. Study on the Biosynthesis of l-SLR by Efficient Prokaryotic Expression of Berberine Bridge Enzyme [J]. Biotechnology Bulletin, 2023, 39(7): 277-287. |
| [2] | CHEN Quan-bing, CAO Wei-jie, LI Chun, LV Bo. Molecular Evolutionary Relationship and Protein Structure of Glycoside Hydrolases from GH79 Family [J]. Biotechnology Bulletin, 2023, 39(1): 104-114. |
| [3] | SUO Qing-qing, WU Nan, YANG Hui, LI Li, WANG Xi-feng. Prokaryotic Expression,Antibody Preparation and Application of Rice Caffeoyl Coenzyme A-O-methyltransferase Gene [J]. Biotechnology Bulletin, 2022, 38(8): 135-141. |
| [4] | QIN Xue-jing, WANG Yu-han, CAO Yi-bo, ZHANG Ling-yun. Prokaryotic Expression and Preparation of Polyclonal Antibody of PwHAP5 Gene in Picea wilsonii [J]. Biotechnology Bulletin, 2022, 38(8): 142-149. |
| [5] | WANG Guang-li, FAN Chan, WANG Hui, LU Hui-fang, XIA Ling-yin, HUANG Jian, MIN Xun. Prokaryotic Expression,Purification,Identification,and Polyclonal Antibody Preparation of Vibrio cholerae Hemolysin HlyA [J]. Biotechnology Bulletin, 2022, 38(7): 269-277. |
| [6] | WANG Qiao-ju, HU Yu-meng, WEN Ya-ya, SONG Li, MENG Chuang, PAN Zhi-ming, JIAO Xin-an. Expression and Activity Identification of SARS-CoV-2 S1 Protein [J]. Biotechnology Bulletin, 2022, 38(3): 157-163. |
| [7] | SHEN Jun-qiang, ZHANG Li-ping, YU Rui-ming, WANG Yong-lu, PAN Li, LIU Xia, LIU Xin-sheng. Porcine Kobuvirus Structural Proteins VP0 and VP1 Prokaryotic Expression and Establishment of Indirect ELISA Method [J]. Biotechnology Bulletin, 2022, 38(10): 243-253. |
| [8] | SHAN Cao-mei, YE Lei, ZHANG Lian-hu, KUANG Wei-gang, SUN Xiao-tang, MA Jian, CUI Ru-qiang. Cloning,and Functional Analysis of Gene OsRAI1 Resistant to Hirschmanniella mucronate in Rice [J]. Biotechnology Bulletin, 2021, 37(7): 146-155. |
| [9] | ZENG Fu-yuan, SU Ze-hui, ZHOU Shi-hui, XIE Miao, PANG Huan-ying. Prokaryotic Expression of the PEPCK Protein of Vibrio alginolyticus and Identification of Its Acetylation and Succinylation [J]. Biotechnology Bulletin, 2021, 37(5): 84-91. |
| [10] | ZHANG Xi-xi, ZHANG Yi-qing, LI Yu-lin, HAN Xiao, WANG Guo-qiang, WANG Xiao-jun, WANG Xu-dong, WANG Yun-long. Prokaryotic Expression,Purification and Application of N Protein C-terminal Recombinant Protein in Novel Coronavirus(SARS-CoV-2) [J]. Biotechnology Bulletin, 2021, 37(5): 92-97. |
| [11] | QU Huan, LI Cheng, CHEN Rui, LIAO Yi-jie, CAO San-jie, WEN Yi-ping, YAN Qi-gui, HUANG Xiao-bo. Truncated Expression of the S1-CTD Fragment of Porcine Deltacoronavirus and Establishment of an Indirect ELISA for Detecting Its Antibody [J]. Biotechnology Bulletin, 2021, 37(5): 273-280. |
| [12] | PENG Li-zhong, ZHANG Peng, ZHOU Wen-wen, ZENG Xu-hui, ZHANG Xiao-ning. Preparation and Multi-purpose Validation of Sperm-specific Protein Cabs1 Polyclonal Antibody [J]. Biotechnology Bulletin, 2021, 37(3): 261-270. |
| [13] | HE Yang, YU Qiao-ling, WANG Jun, QIN Chuan-jie, LI Hua-tao. Advances in Prokaryotic Expression Gene of Tilapia [J]. Biotechnology Bulletin, 2021, 37(2): 195-202. |
| [14] | TANG Lu, DONG Li-ping, YIN Mo-li, LIU Lei, DONG Yuan, WANG Hui-yan. Preparation and Identification of a Novel FGF20 Monoclonal Antibody [J]. Biotechnology Bulletin, 2021, 37(10): 179-185. |
| [15] | DUAN Ying-ce, HU Zi-yi, YANG Fan, LI Jin-tao, WU Xiang-li, ZHANG Rui-ying. Cloning and Expression Analysis of Oxaloacetate Hydrolase(LeOAH1)Gene from Lentinula edodes [J]. Biotechnology Bulletin, 2020, 36(9): 227-234. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||