








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (6): 136-146.doi: 10.13560/j.cnki.biotech.bull.1985.2020-1385
Previous Articles Next Articles
HUANG Jing-xiao( ), SHANG Jun-kang, CHEN Hui-min, SHEN Jia-min, LI Yuan-yuan, YU Yu-li, NI Jin-dong(
), SHANG Jun-kang, CHEN Hui-min, SHEN Jia-min, LI Yuan-yuan, YU Yu-li, NI Jin-dong( ), LIN Bo-kun(
), LIN Bo-kun( )
)
Received:2020-11-13
Online:2021-06-26
Published:2021-07-08
Contact:
NI Jin-dong,LIN Bo-kun
E-mail:1562895101@qq.com;nijd-gw@gdmu.edu.cn;bklin@gdmu.edu.cn
HUANG Jing-xiao, SHANG Jun-kang, CHEN Hui-min, SHEN Jia-min, LI Yuan-yuan, YU Yu-li, NI Jin-dong, LIN Bo-kun. Biological Characterization and Genome Analysis of a Lytic Phage Infecting Salmonella[J]. Biotechnology Bulletin, 2021, 37(6): 136-146.
| 宿主菌 Host bacterium | 宿主菌编号 Host No. | PSM6裂解活性 Infectivity of PSM6 |
|---|---|---|
| Salmonella Typhimurium | J25、J29、J45、J47、J49 | ++ |
| Salmonella Derby | J07 | ++ |
| Salmonella Derby | J23 | + |
| Salmonella Rissen | J13、J14、J20、J26 | - |
| Salmonella Corvallis | J15、J30 | ++ |
| Salmonella Acina | J18 | - |
| Salmonella London | J19、J24 | ++ |
| Salmonella Agona | J22、J58 | - |
| Escherichia coli | ATCC25922、DH5α、BL21 | ++ |
| Shigella Sonnei | CMCC(B)51592 | + |
| Shigella Flexneri | CMCC(B)51572 | - |
| Shigella Dysenteriae | CMCC(B)51105 | - |
| Shigella | ATCC14029 | - |
Table 1 Host range of phages PSM6
| 宿主菌 Host bacterium | 宿主菌编号 Host No. | PSM6裂解活性 Infectivity of PSM6 |
|---|---|---|
| Salmonella Typhimurium | J25、J29、J45、J47、J49 | ++ |
| Salmonella Derby | J07 | ++ |
| Salmonella Derby | J23 | + |
| Salmonella Rissen | J13、J14、J20、J26 | - |
| Salmonella Corvallis | J15、J30 | ++ |
| Salmonella Acina | J18 | - |
| Salmonella London | J19、J24 | ++ |
| Salmonella Agona | J22、J58 | - |
| Escherichia coli | ATCC25922、DH5α、BL21 | ++ |
| Shigella Sonnei | CMCC(B)51592 | + |
| Shigella Flexneri | CMCC(B)51572 | - |
| Shigella Dysenteriae | CMCC(B)51105 | - |
| Shigella | ATCC14029 | - |
| 感染 复数 MOI | 噬菌体效价 Titer of phage (PFU·mL-1) | 宿主菌浓度 Concentration of host bacteria (CFU·mL-1) | 培养3 h后的噬菌体效价 The titer of phage after 3 h culture (PFU·mL-1) |
|---|---|---|---|
| 100 | 1.0×105 | 1.0×103 | (9.4±0.2)×108 |
| 10 | 1.0×105 | 1.0×104 | (2.2±0.5)×108 |
| 1 | 1.0×105 | 1.0×105 | (6.2±0.6)×107 |
| 0.1 | 1.0×105 | 1.0×106 | (8.1±0.5)×108 |
| 0.01 | 1.0×105 | 1.0×107 | (1.2±0.6)×1010 |
| 0.001 | 1.0×105 | 1.0×108 | (4.7±0.3)×109 |
Table 2 Optimal multiplicity of infection(MOI)of phages PSM6
| 感染 复数 MOI | 噬菌体效价 Titer of phage (PFU·mL-1) | 宿主菌浓度 Concentration of host bacteria (CFU·mL-1) | 培养3 h后的噬菌体效价 The titer of phage after 3 h culture (PFU·mL-1) |
|---|---|---|---|
| 100 | 1.0×105 | 1.0×103 | (9.4±0.2)×108 |
| 10 | 1.0×105 | 1.0×104 | (2.2±0.5)×108 |
| 1 | 1.0×105 | 1.0×105 | (6.2±0.6)×107 |
| 0.1 | 1.0×105 | 1.0×106 | (8.1±0.5)×108 |
| 0.01 | 1.0×105 | 1.0×107 | (1.2±0.6)×1010 |
| 0.001 | 1.0×105 | 1.0×108 | (4.7±0.3)×109 |
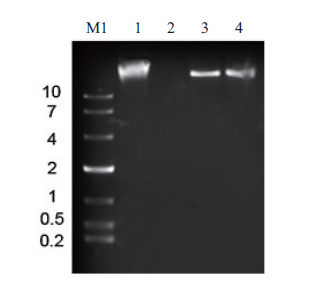
Fig.6 Nucleic acid type of phage PSM6 M1:10 Kb DNA molecular weight standard. 1:PSM6 nucleic acid. 2:DNase I PSM6 nucleic acid. 3:Mung-Bean Nuclease PSM6 nucleic acid. 4:RNase A digests PSM6 nucleic acid
| 噬菌体名称 Phage name | 大小Genome length/bp | GC含量 GC content/% | 编码区 Coding region | 编码区平均大小 Coding region length/bp | 编码区占百分比 Encoding area percent/% | 转运RNA Transfer RNA | GenBank登录号 Accession No. |
|---|---|---|---|---|---|---|---|
| PSM6 | 90730 | 39.57% | 133 | 601.8 | 88.2 | 25 | This study |
| Salmonella phage vB_SPuM_SP116 | 87510 | 38.29% | 126 | 604.1 | 87.0 | 22 | KP010413.1 |
| Escherichia phage vB_EcoM_AYO145A | 87372 | 39.00% | 131 | 592.6 | 88.9 | 20 | KR014248.1 |
| Escherichia phage VpaE1_ev108 | 87749 | 38.92% | 129 | 596.1 | 87.6 | 26 | LR597658.1 |
| Escherichia phage VpaE1_ev035 | 88002 | 38.96% | 132 | 591.4 | 88.0 | 26 | LR699048.1 |
| Escherichia phage NBEco005 | 88272 | 38.84% | 130 | 593.6 | 87.4 | 19 | MN994499.1 |
| Shigella phage Z31 | 89355 | 38.88% | 132 | 594.7 | 87.9 | 26 | MN655999.1 |
| Salmonella phage D1-2 | 86878 | 38.73% | 128 | 577.6 | 85.1 | 23 | MN481367.1 |
Table 3 Comparative analysis of PSM6 and its closely related phage
| 噬菌体名称 Phage name | 大小Genome length/bp | GC含量 GC content/% | 编码区 Coding region | 编码区平均大小 Coding region length/bp | 编码区占百分比 Encoding area percent/% | 转运RNA Transfer RNA | GenBank登录号 Accession No. |
|---|---|---|---|---|---|---|---|
| PSM6 | 90730 | 39.57% | 133 | 601.8 | 88.2 | 25 | This study |
| Salmonella phage vB_SPuM_SP116 | 87510 | 38.29% | 126 | 604.1 | 87.0 | 22 | KP010413.1 |
| Escherichia phage vB_EcoM_AYO145A | 87372 | 39.00% | 131 | 592.6 | 88.9 | 20 | KR014248.1 |
| Escherichia phage VpaE1_ev108 | 87749 | 38.92% | 129 | 596.1 | 87.6 | 26 | LR597658.1 |
| Escherichia phage VpaE1_ev035 | 88002 | 38.96% | 132 | 591.4 | 88.0 | 26 | LR699048.1 |
| Escherichia phage NBEco005 | 88272 | 38.84% | 130 | 593.6 | 87.4 | 19 | MN994499.1 |
| Shigella phage Z31 | 89355 | 38.88% | 132 | 594.7 | 87.9 | 26 | MN655999.1 |
| Salmonella phage D1-2 | 86878 | 38.73% | 128 | 577.6 | 85.1 | 23 | MN481367.1 |
| 蛋白编号 Protein No. | 蛋白功能预测 Protein function prediction | 蛋白编号 Protein No. | 蛋白功能预测 Protein function prediction | |
|---|---|---|---|---|
| GENE8 | D-alanyl-D-alanine carboxypeptidase | GENE65 | Minor tail protein | |
| GENE10 | Holin | GENE66 | Deoxynucleotide monophosphate kinase | |
| GENE11 | Tail protein | GENE68 | DNA helicase activity | |
| GENE17 | Lysin | GENE71 | Exodeoxyribonuclease | |
| GENE18 | Putative membrane protein | GENE72 | NAD synthetase | |
| GENE20 | Terminase | GENE74 | Protein disulfide oxidoreductase | |
| GENE21 | Structural protein | GENE77 | Ribonucleotide reductase | |
| GENE24 | Peptidase family S49 | GENE79 | Ribonucleotide reductase | |
| GENE25 | Structural protein | GENE80 | Protein disulfide oxidoreductase | |
| GENE26 | Phage major capsid protein E | GENE84 | Ribonucleoside-triphosphate reductase | |
| GENE31 | Structural protein | GENE86 | Ribonucleoside-triphosphate reductase-activating protein | |
| GENE33 | Tape measure chaperone | GENE92 | Ribose-phosphate pyrophosphokinase | |
| GENE35 | Tape measure chaperone | GENE93 | Nicotinate-nucleotide diphosphorylase | |
| GENE39 | Baseplate assembly protein | GENE106 | rIIa protein | |
| GENE41 | Baseplate assembly protein | GENE107 | rIIB lysis inhibitor | |
| GENE45 | Tail fiber protein | GENE108 | Lysin,lytic transglycosylase | |
| GENE47 | Holin | GENE109 | Polynucleotide kinase | |
| GENE48 | Thymidylate synthase | GENE111 | i-spannin | |
| GENE49 | Dihydrofolate reductase | GENE113 | O-spanin | |
| GENE53 | Transcriptional regulatory protein | GENE114 | Tail assembly protein | |
| GENE56 | DNA ligase | GENE117 | Phosphatase | |
| GENE61 | DNA polymerase family A | GENE123 | Tail sheath monomer | |
| GENE62 | Endonuclease activity | GENE124 | Tail tube protein | |
| GENE63 | DNA polymerase family A |
Table 4 Prediction of PSM6 protein-coding regions
| 蛋白编号 Protein No. | 蛋白功能预测 Protein function prediction | 蛋白编号 Protein No. | 蛋白功能预测 Protein function prediction | |
|---|---|---|---|---|
| GENE8 | D-alanyl-D-alanine carboxypeptidase | GENE65 | Minor tail protein | |
| GENE10 | Holin | GENE66 | Deoxynucleotide monophosphate kinase | |
| GENE11 | Tail protein | GENE68 | DNA helicase activity | |
| GENE17 | Lysin | GENE71 | Exodeoxyribonuclease | |
| GENE18 | Putative membrane protein | GENE72 | NAD synthetase | |
| GENE20 | Terminase | GENE74 | Protein disulfide oxidoreductase | |
| GENE21 | Structural protein | GENE77 | Ribonucleotide reductase | |
| GENE24 | Peptidase family S49 | GENE79 | Ribonucleotide reductase | |
| GENE25 | Structural protein | GENE80 | Protein disulfide oxidoreductase | |
| GENE26 | Phage major capsid protein E | GENE84 | Ribonucleoside-triphosphate reductase | |
| GENE31 | Structural protein | GENE86 | Ribonucleoside-triphosphate reductase-activating protein | |
| GENE33 | Tape measure chaperone | GENE92 | Ribose-phosphate pyrophosphokinase | |
| GENE35 | Tape measure chaperone | GENE93 | Nicotinate-nucleotide diphosphorylase | |
| GENE39 | Baseplate assembly protein | GENE106 | rIIa protein | |
| GENE41 | Baseplate assembly protein | GENE107 | rIIB lysis inhibitor | |
| GENE45 | Tail fiber protein | GENE108 | Lysin,lytic transglycosylase | |
| GENE47 | Holin | GENE109 | Polynucleotide kinase | |
| GENE48 | Thymidylate synthase | GENE111 | i-spannin | |
| GENE49 | Dihydrofolate reductase | GENE113 | O-spanin | |
| GENE53 | Transcriptional regulatory protein | GENE114 | Tail assembly protein | |
| GENE56 | DNA ligase | GENE117 | Phosphatase | |
| GENE61 | DNA polymerase family A | GENE123 | Tail sheath monomer | |
| GENE62 | Endonuclease activity | GENE124 | Tail tube protein | |
| GENE63 | DNA polymerase family A |
| [1] |
Aliakbar Ahovan Z, Hashemi A, de Plano LM, et al. Bacteriophage based biosensors:trends, outcomes and challenges[J]. Nanomaterials, 2020, 10(3):501.
doi: 10.3390/nano10030501 URL |
| [2] |
Li SS, Liu S, Xu YC, et al. Robust and highly specific fluorescence sensing of Salmonella typhimurium based on dual-functional phi29 DNA polymerase-mediated isothermal circular strand displacement polymerization[J]. Analyst, 2019, 144(16):4795-4802.
doi: 10.1039/C9AN00843H URL |
| [3] |
Kirk MD, Pires SM, Black RE, et al. Correction:World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010:A data synjournal[J]. PLoS Med, 2015, 12(12):e1001940.
doi: 10.1371/journal.pmed.1001940 URL |
| [4] |
Yousefi H, Ali MM, Su HM, et al. Sentinel wraps:real-time monitoring of food contamination by printing DNAzyme probes on food packaging[J]. ACS Nano, 2018, 12(4):3287-3294.
doi: 10.1021/acsnano.7b08010 pmid: 29621883 |
| [5] |
GBD Non-Typhoidal Salmonella Invasive Disease Collaborators. The global burden of non-typhoidal salmonella invasive disease:a systematic analysis for the global burden of disease study 2017[J]. Lancet Infect Dis, 2019, 19(12):1312-1324.
doi: 10.1016/S1473-3099(19)30418-9 URL |
| [6] |
GBD 2017 Typhoid and Paratyphoid Collaborators. The global burden of typhoid and paratyphoid fevers:a systematic analysis for the global burden of disease study 2017[J]. Lancet Infect Dis, 2019, 19(4):369-381.
doi: 10.1016/S1473-3099(18)30685-6 URL |
| [7] |
Merino L, Procura F, Trejo FM, et al. Biofilm formation by Salmonella sp. in the poultry industry:detection, control and eradication strategies[J]. Food Res Int, 2019, 119:530-540.
doi: S0963-9969(17)30788-3 pmid: 30884686 |
| [8] |
Martínez MC, Retamal P, et al. Multidrug-resistant outbreak-associated Salmonella strains in irrigation water from the metropolitan reg-ion, Chile[J]. Zoonoses Public Health, 2017, 64(4):299-304.
doi: 10.1111/zph.2017.64.issue-4 URL |
| [9] |
Lee D, Tertuliano M, Harris C, et al. Salmonella survival in soil and transfer onto produce via splash events[J]. J Food Prot, 2019, 82(12):2023-2037.
doi: 10.4315/0362-028X.JFP-19-066 URL |
| [10] |
Biasino W, de Zutter L, Mattheus W, et al. Correlation between slaughter practices and the distribution of Salmonella and hygiene indicator bacteria on pig carcasses during slaughter[J]. Food Microbiol, 2018, 70:192-199.
doi: S0740-0020(17)30641-X pmid: 29173627 |
| [11] |
Lamas A, Regal P, Vázquez B, et al. Salmonella and Campylobacter biofilm formation:a comparative assessment from farm to fork[J]. J Sci Food Agric, 2018, 98(11):4014-4032.
doi: 10.1002/jsfa.2018.98.issue-11 URL |
| [12] |
Xu Y, Tao S, Hinkle N, et al. Salmonella, including antibiotic-resistant Salmonella, from flies captured from cattle farms in Georgia, U. S. A[J]. Sci Total Environ, 2018, 616-617:90-96.
doi: 10.1016/j.scitotenv.2017.10.324 URL |
| [13] | Kuehn B. Multidrug-resistant Salmonella[J]. JAMA, 2019, 322(14):1344. |
| [14] | Hawkey J, Le HelloS, Doublet B, et al. Global phylogenomics of multidrug-resistant Salmonella enterica serotype Kentucky ST198[J]. Microb Genom, 2019, 5(7):e000269. |
| [15] |
Váradi L, Luo JL, Hibbs DE, et al. Methods for the detection and identification of pathogenic bacteria:past, present, and future[J]. Chem Soc Rev, 2017, 46(16):4818-4832.
doi: 10.1039/c6cs00693k pmid: 28644499 |
| [16] |
Carvalho C, Costa AR, Silva F, et al. Bacteriophages and their derivatives for the treatment and control of food-producing animal infections[J]. Crit Rev Microbiol, 2017, 43(5):583-601.
doi: 10.1080/1040841X.2016.1271309 pmid: 28071145 |
| [17] |
Wei S, Chelliah R, Rubab M, et al. Bacteriophages as potential tools for detection and control of Salmonella spp. in food systems[J]. Microorganisms, 2019, 7(11):570.
doi: 10.3390/microorganisms7110570 URL |
| [18] |
Oh JH, Park MK . Recent trends in Salmonella outbreaks and emerging technology for biocontrol of Salmonella using phages in foods:a review[J]. J Microbiol Biotechnol, 2017, 27(12):2075-2088.
doi: 10.4014/jmb.1710.10049 URL |
| [19] |
Kawacka I, Olejnik-Schmidt A, et al. Effectiveness of phage-based inhibition of Listeria monocytogenes in food products and food proc-essing environments[J]. Microorganisms. 2020, 8(11):1764.
doi: 10.3390/microorganisms8111764 URL |
| [20] | Rehman S, Ali Z, Khan M, et al. The dawn of phage therapy[J]. Rev Med Virol, 2019, 29(4):e2041. |
| [21] | 许燕苹. 沙门菌噬菌体的分离鉴定及在禽肉产品中的初步应用[D]. 扬州:扬州大学, 2018. |
| Xu YP. The isolation and identification of Salmonella bacteriophages and their preliminary application on poultry meats[D]. Yangzhou:Yangzhou University, 2018. | |
| [22] |
Huang C, Virk SM, Shi J, et al. Isolation, characterization, and application of bacteriophage LPSE1 against Salmonella enterica in ready to eat(RTE)foods[J]. Front Microbiol, 2018, 9:1046.
doi: 10.3389/fmicb.2018.01046 URL |
| [23] |
Phothaworn P, Supokaivanich R, Lim J, et al. Development of a broad-spectrum Salmonella phage cocktail containing Viunalike and Jerseylike viruses isolated from Thailand[J]. Food Microbiol, 2020, 92:103586.
doi: S0740-0020(20)30175-1 pmid: 32950171 |
| [24] |
Endersen L, O’Mahony J, et al. Phage therapy in the food industry[J]. Annu Rev Food Sci Technol, 2014, 5(1):327-349.
doi: 10.1146/annurev-food-030713-092415 URL |
| [25] |
Li Y, Yang X, Zhang H, et al. Prevalence and antimicrobial susceptibility of Salmonella in the commercial eggs in China[J]. Int J Food Microbiol, 2020, 325:108623.
doi: 10.1016/j.ijfoodmicro.2020.108623 URL |
| [26] | European Food Safety Authority and European Centre for Disease Prevention and Control. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017[J]. EFSA J, 2018, 16(12):e5500. |
| [27] |
Shaw KA, Wright K, Privett K, et al. Salmonellosis outbreak after a large-scale food event in Virginia, 2017[J]. Public Health Rep, 2020, 135(5):668-675.
doi: 10.1177/0033354920944861 URL |
| [28] |
Wang X, Sun J, Zhao J, et al. All-solid-state fiber-shaped asymmetric supercapacitors with ultrahigh energy density based on porous vanadium nitride nanowires and ultrathin Ni(OH)2 nanosheet wrapped NiCo2O4 nanowires arrays electrode[J]. The Journal of Physical Chemistry C, 2019, 123(2):985-993.
doi: 10.1021/acs.jpcc.8b05862 URL |
| [29] |
Latash J, Greene SK, Stavinsky F, et al. Salmonellosis outbreak detected by automated spatiotemporal analysis-New York city, May-June 2019[J]. MMWR Morb Mortal Wkly Rep, 2020, 69(26):815-819.
doi: 10.15585/mmwr.mm6926a2 URL |
| [30] |
Maio R, García-Díez J, Saraiva C. Microbiological quality of foodstuffs sold on Expiry date at retail in portugal:a preliminary study[J]. Foods, 2020, 9(7):919.
doi: 10.3390/foods9070919 URL |
| [31] |
Ren D, Chen P, et al. Phenotypes and antimicrobial resistance genes in Salmonella isolated from retail chicken and pork in Changchun, China[J]. J Food Safety, 2017, 37(2):e12314.
doi: 10.1111/jfs.2017.37.issue-2 URL |
| [32] | Ramatla TA, Mphuthi N, Ramaili T, et al. Molecular detection of virulence genes in Salmonella spp. isolated from chicken faeces in Mafikeng, South Africa[J]. J S Afr Vet Assoc, 2020, 91:a1994. |
| [33] |
Lal TM, Sano M, Ransangan J. Isolation and characterization of large marine bacteriophage(Myoviridae), VhKM4 infecting vibrio harveyi[J]. J Aquat Anim Health, 2017, 29(1):26-30.
doi: 10.1080/08997659.2016.1249578 URL |
| [34] |
Wongsuntornpoj S, Moreno Switt AI, Bergholz P, et al. Salmonella phages isolated from dairy farms in Thailand show wider host range than a comparable set of phages isolated from US dairy farms[J]. Vet Microbiol, 2014, 172:345-352.
doi: 10.1016/j.vetmic.2014.05.023 pmid: 24939592 |
| [35] |
Huang C, Shi J, Ma W, et al. Isolation, characterization, and application of a novel specific Salmonella bacteriophage in different food matrices[J]. Food Res Int, 2018, 111:631-641.
doi: 10.1016/j.foodres.2018.05.071 URL |
| [36] | 王孝芳, 侯玉刚, 等. 一株青枯菌专性噬菌体的分离及应用效果研究[J]. 生物技术通报, 2020, 36(9):194-201. |
| Wang XF, Hou YG, et al. Isolation of specific phage of Ralstonia solanacearum and its effects on control of soil-borne bacterial wilt disease[J]. Biotechnology Bulletin, 2020, 36(9):194-201. | |
| [37] |
Kowalska JD, Kazimierczak J, Sowińska PM, et al. Growing trend of fighting infections in aquaculture environment- opportunities and challenges of phage therapy[J]. Antibiotics, 2020, 9(6):301.
doi: 10.3390/antibiotics9060301 URL |
| [38] |
Imklin N, Nasanit R. Characterization of Salmonella bacteriophages and their potential use in dishwashing materials[J]. J Appl Microbiol, 2020, 129(2):266-277.
doi: 10.1111/jam.14617 pmid: 32073713 |
| [39] |
Bao H, Zhang P, Zhang H, et al. Bio-control of Salmonella Enteritidis in foods using bacteriophages[J]. Viruses, 2015, 7(8):4836-4853.
doi: 10.3390/v7082847 URL |
| [40] |
Buttimer C, Lynch C, et al. Isolation and characterization of Pectobacterium Phage vB_PatM_CB7:new insights into the genus Certrevirus[J]. Antibiotics, 2020, 9(6):352.
doi: 10.3390/antibiotics9060352 URL |
| [41] |
Liu J, Gao S, Dong Y, et al. Isolation and characterization of bacteriophages against virulent Aeromonas hydrophila[J]. BMC Microbiol, 2020, 20(1):141.
doi: 10.1186/s12866-020-01811-w URL |
| [42] |
Kim HJ, Giri SS, Kim SG, et al. Isolation and characterization of two bacteriophages and their preventive effects against pathogenic Vibrio coralliilyticus causing mortality of pacific oyster(crassostrea gigas)larvae[J]. Microorganisms, 2020, 8(6):926.
doi: 10.3390/microorganisms8060926 URL |
| [43] |
Yan T, Liang L, Yin P, et al. Application of a novel phage LPSEYT for biological control of Salmonella in foods[J]. Microorganisms, 2020, 8(3):400.
doi: 10.3390/microorganisms8030400 URL |
| [44] |
Phothaworn P, Dunne M, Supokaivanich R, et al. Characterization of flagellotropic, chi-Like Salmonella phages isolated from Thai poultry farms[J]. Viruses, 2019, 11(6):520.
doi: 10.3390/v11060520 URL |
| [45] |
Islam MS, Hu Y, Mizan MFR, et al. Characterization of Salmonella phage LPST153 that effectively targets most prevalent Salmonella Serovars[J]. Microorganisms, 2020, 8(7):1089.
doi: 10.3390/microorganisms8071089 URL |
| [46] |
Dion MB, Oechslin F, Moineau S. Phage diversity, genomics and phylogeny[J]. Nat Rev Microbiol, 2020, 18(3):125-138.
doi: 10.1038/s41579-019-0311-5 URL |
| [47] | 冯烨, 刘军, 孙洋, 等. 噬菌体最新分类与命名[J]. 中国兽医学报, 2013, 33(12):1954-1958. |
| Feng Y, Liu J, Sun Y, et al. An introduction to current classification and nomenclature of bacterial viruses[J]. Chinese Journal of Veterinary Science, 2013, 33(12):1954-1958. | |
| [48] |
Lefkowitz EJ, Dempsey DM, Hendrickson RC, et al. Virus taxonomy:the database of the International Committee on Taxonomy of Viruses(ICTV)[J]. Nucleic Acids Res, 2018, 46:D708-D717.
doi: 10.1093/nar/gkx932 URL |
| [49] | 王宁宁, 魏晓宇, 尚津宇, 等. 噬菌体治疗细菌感染的机制及现状[J]. 吉林医药学院学报, 2019, 40(6):437-439. |
| Wang NN, Wei XY, Shang JY, et al. The mechanism of phage therapy for bacterial infection and its status[J]. Journal of Jilin Medical University, 2019, 40(6):437-439. | |
| [50] |
Bao H, Shahin K, Zhang Q, et al. Morphologic and genomic characterization of a broad host range Salmonella enterica serovar pullorum lytic phage vB_SPuM_SP116[J]. Microb Pathog, 2019, 136:103659.
doi: 10.1016/j.micpath.2019.103659 URL |
| [51] | Bardina C, Colom J, Spricigo DA, et al. Genomics of three new bacteriophages useful in the biocontrol of Salmonella[J]. Front Microbiol, 2016, 7:545. |
| [52] |
Vila J, Moreno-Morales J, Ballesté-Delpierre C. Current landscape in the discovery of novel antibacterial agents[J]. Clin Microbiol Infect, 2020, 26(5):596-603.
doi: 10.1016/j.cmi.2019.09.015 URL |
| [53] |
Wernicki A, Nowaczek A, et al. Bacteriophage therapy to combat bacterial infections in poultry[J]. Virol J, 2017, 14(1):179.
doi: 10.1186/s12985-017-0849-7 pmid: 28915819 |
| [1] | ZHANG Kun, YAN Chang, TIAN Xin-peng. Research Progress in Microbial Single Cell Separation Methods [J]. Biotechnology Bulletin, 2023, 39(9): 1-11. |
| [2] | WANG Teng-hui, GE Wen-dong, LUO Ya-fang, FAN Zhen-yu, WANG Yu-shu. Gene Mapping of Kale White Leaves Based on Whole Genome Re-sequencing of Extreme Mixed Pool(BSA) [J]. Biotechnology Bulletin, 2023, 39(9): 176-182. |
| [3] | LI Xue-qi, ZHANG Su-jie, YU Man, HUANG Jin-guang, ZHOU Huan-bin. Establishment of CRISPR/CasX-based Genome Editing Technology in Rice [J]. Biotechnology Bulletin, 2023, 39(9): 40-48. |
| [4] | FANG Lan, LI Yan-yan, JIANG Jian-wei, CHENG Sheng, SUN Zheng-xiang, ZHOU Yi. Isolation, Identification and Growth-promoting Characteristics of Endohyphal Bacterium 7-2H from Endophytic Fungi of Spiranthes sinensis [J]. Biotechnology Bulletin, 2023, 39(8): 272-282. |
| [5] | RAO Zi-huan, XIE Zhi-xiong. Isolation and Identification of a Cellulose-degrading Strain of Olivibacter jilunii and Analysis of Its Degradability [J]. Biotechnology Bulletin, 2023, 39(8): 283-290. |
| [6] | GUO Shao-hua, MAO Hui-li, LIU Zheng-quan, FU Mei-yuan, ZHAO Ping-yuan, MA Wen-bo, LI Xu-dong, GUAN Jian-yi. Whole Genome Sequencing and Comparative Genome Analysis of a Fish-derived Pathogenic Aeromonas Hydrophila Strain XDMG [J]. Biotechnology Bulletin, 2023, 39(8): 291-306. |
| [7] | ZHANG Dao-lei, GAN Yu-jun, LE Liang, PU Li. Epigenetic Regulation of Yield-related Traits in Maize and Epibreeding [J]. Biotechnology Bulletin, 2023, 39(8): 31-42. |
| [8] | SHI Jia-xin, LIU Kai, ZHU Jin-jie, QI Xian-tao, XIE Chuan-xiao, LIU Chang-lin. Gene Editing Reshaping Maize Plant Type for Increasing Hybrid Yield [J]. Biotechnology Bulletin, 2023, 39(8): 62-69. |
| [9] | DU Dong-dong, QIAN Jing, LI Si-qi, LIU Wen-fei, WEI Xiang-li, LIU Chang-yong, LUO Rui-feng, KANG Li-chao. Whole Genome Sequencing and Analysis of Listeria monocytogenes Strain LMXJ15 [J]. Biotechnology Bulletin, 2023, 39(7): 298-306. |
| [10] | LI Yu-zhen, MEI Tian-xiu, LI Zhi-wen, WANG Qi, LI Jun, ZOU Yue, ZHAO Xin-qing. Advances in Genomic Studies and Metabolic Engineering of Red Yeasts [J]. Biotechnology Bulletin, 2023, 39(7): 67-79. |
| [11] | YOU Zi-juan, CHEN Han-lin, DENG Fu-cai. Research Progress in the Extraction and Functional Activities of Bioactive Peptides from Fish Skin [J]. Biotechnology Bulletin, 2023, 39(7): 91-104. |
| [12] | YIN Ming-hua, YU Huan-yuan, XIAO Xin-yi, WANG Yu-ting. Chloroplast Genomic Characterization and Phylogenetic Analysis of Colocasia esculenta L. Schoot var. cormosus cv. ‘Hongyayu’ from Jiangxi Yanshan [J]. Biotechnology Bulletin, 2023, 39(6): 233-247. |
| [13] | ZHANG Lu-yang, HAN Wen-long, XU Xiao-wen, YAO Jian, LI Fang-fang, TIAN Xiao-yuan, ZHANG Zhi-qiang. Identification and Expression Analysis of the Tobacco TCP Gene Family [J]. Biotechnology Bulletin, 2023, 39(6): 248-258. |
| [14] | LI Tuo, LI Long-ping, QU Lei. Research Progress in the Structure of Tailed Bacteriophage and Its Receptors [J]. Biotechnology Bulletin, 2023, 39(6): 88-101. |
| [15] | CHE Yong-mei, GUO Yan-ping, LIU Guang-chao, YE Qing, LI Ya-hua, ZHAO Fang-gui, LIU Xin. Isolation and Identification of Bacterial Strain C8 and B4 and Their Halotolerant Growth-promoting Effects and Mechanisms [J]. Biotechnology Bulletin, 2023, 39(5): 276-285. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||